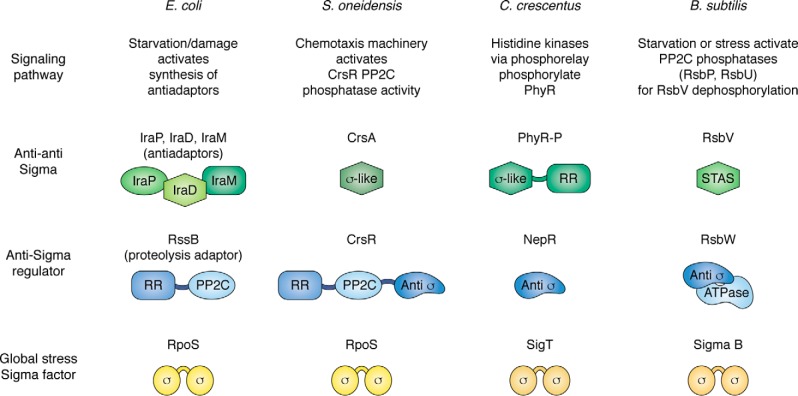
Figure 5.
Anti-sigmas and anti-anti–sigmas in general stress responses. On this chart, sigma factors mediating general stress responses are shown in shades of yellow and brown. Alphaproteobacteria use an ECF family sigma factor, named SigT in C. crescentus, shown here, and are referred to more generally as σEcfG in other alphaproteobacteria, to mediate their general stress response. RpoS (E. coli and S. oneidensis) and sigma B (in B. subtilis and some other Gram-positive bacteria) are related to the vegetative sigma factors. Blue symbols show general organization of the proteins that operate as “anti-sigmas,” although RssB, as noted, targets RpoS for proteolysis rather than simply binding it, as for other anti-sigmas. RR domains are response regulators. The PP2C domain in RssB is inactive as a phosphatase (pale blue oval). Some of the anti-anti–sigmas have sigma-like domains, providing mimics for binding to the anti-sigmas. STAS stands for sulfate transporter and anti-sigma factor antagonist domain; CrsA is also a STAS protein. PP2C domains (ovals in anti-sigmas) are also found in the upstream signaling pathways of many of these organisms (for more detail see Ref. 117). Not shown here is the signaling pathway upstream of PhyR–P, in which multiple histidine kinases converge on response regulator MrrA, which in turns transfers phosphate via PhyK to PhyR (131).