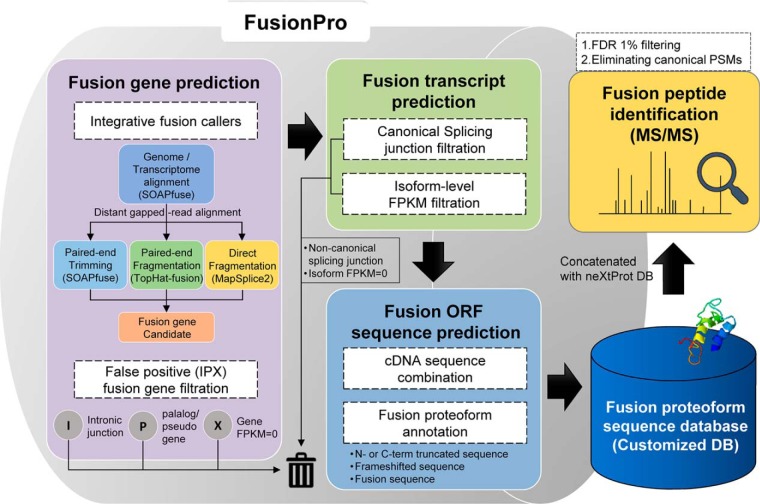
Fig. 1.
Data analysis workflow of FusionPro. FusionPro contains modules that provide predictions of the fusion transcript and fusion proteoform sequences. First, the gene fusion prediction module provides a list of candidate fusion transcripts annotated by selective integration of the workflow of the three fusion finders (SOAPfuse, TopHat-fusion, and MapSplice2). From the predicted fusion genes or user-specified fusion genes, the “IPX” filtration module filters out the false positives and integrates the results into a single data format. Finally, the fusion cDNA sequences are three-frame translated and annotated to construct a customized DB for the identification of the fusion proteoforms.