Figure 10.
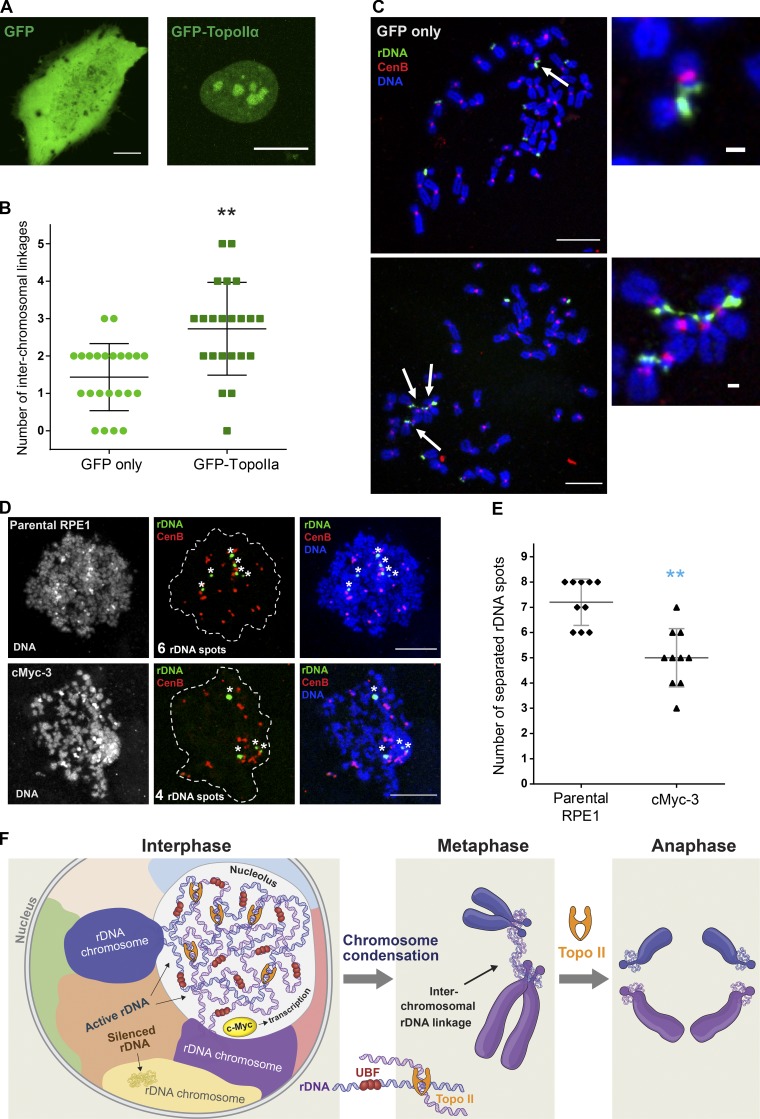
Topoisomerase IIα plays a major role in creating and resolving rDNA linkages. (A) Fluorescent images of interphase cells transiently transfected for 24 h with GFP-topoisomerase IIα (TopoIIα; top) or GFP only (bottom) are shown. GFP-TopoIIα shows propensity to localize in nucleoli. Bar, 10 µm. (B) The number of interchromosomal rDNA linkages in chromosomal spreads from cells transiently transfected with GFP or GFP-TopoIIα was quantified. For this experiment, RPE1 cells were transfected with indicated plasmids for 24 h and arrested in mitosis by addition of colcemid for 12–14 h. Mitotic cells were collected by shake-off and FACS sorted to isolate GFP-positive mitotic cells. Mitotic spreads from cells expressing GFP-TopoIIα show significantly more rDNA linkages compared with GFP only. The Mann–Whitney U test was used to compare GFP-TopoIIα–expressing samples with GFP-expressing control. **, P = 0.0002. (C) Representative confocal images of chromosome spreads from cells transiently transfected for with GFP-TopoIIα, GFP only (top), or TopoIIα (bottom). Spreads were labeled by FISH with rDNA probe (green) and CenB probe (red). Bar, 10 µm. Arrows point to rDNA linkages shown in magnified insets on the right; bar, 1 µm. (D) Chromatin spreads from interphase parental RPE1 cells (top) and cMyc-3 derivative (bottom) cells. Interphase cultures were treated with 100 nM calyculin A for 90 min to induce premature chromatin condensation and labeled by FISH with rDNA probe (green) and CenB probe (red). Asterisks denote individual separated rDNA spots. Bar, 10 µm. (E) Quantification of the number of separated rDNA spots in 100 nM calyculin A–treated interphase cells (D) labeled by FISH with rDNA probe. High-resolution confocal images of ≥10 condensed chromatin spreads were examined. The Mann–Whitney U test was used to compare c-Myc–overexpressing samples with parental RPE1. **, P < 0.001. (F) Working model for how interchromosomal rDNA linkages are generated and resolved. Inter- and intrachromosomal rDNA catenations may form due to the intertwining of transcriptionally active DNA from different chromosomes in the crowded nucleolar environment. Formation of rDNA linkages depends on UBF, and boosting rDNA transcription by overexpressing c-Myc increases the frequency of rDNA linkages. Silent (UBF−) loci are not incorporated in nucleoli and do not form linkages. Topoisomerase II generates more catenations by trying to correct the local topology in the interphase nucleus but then resolves these catenations during the metaphase–anaphase transition. Therefore, transcription-dependent formation of rDNA linkages does not lead to chromosomal missegregation and genomic instability under normal conditions.