The Mycobacterium tuberculosis genome is more heterogenous and less genetically stable within the host than previously thought. Currently, only limited data exist on the within-host microevolution, diversity, and genetic stability of M. tuberculosis.
KEYWORDS: Mycobacterium tuberculosis, drug resistance, evolution, whole-genome sequencing, within-host
SUMMARY
The Mycobacterium tuberculosis genome is more heterogenous and less genetically stable within the host than previously thought. Currently, only limited data exist on the within-host microevolution, diversity, and genetic stability of M. tuberculosis. As a direct consequence, our ability to infer M. tuberculosis transmission chains and to understand the full complexity of drug resistance profiles in individual patients is limited. Furthermore, apart from the acquisition of certain drug resistance-conferring mutations, our knowledge on the function of genetic variants that emerge within a host and their phenotypic impact remains scarce. We performed a systematic literature review of whole-genome sequencing studies of serial and parallel isolates to summarize the knowledge on genetic diversity and within-host microevolution of M. tuberculosis. We identified genomic loci of within-host emerged variants found across multiple studies and determined their functional relevance. We discuss important remaining knowledge gaps and finally make suggestions on the way forward.
INTRODUCTION
For many decades, it was believed that Mycobacterium tuberculosis infections are genetically homogenous and remain stable within the host during infection. With the advent of phage typing and the introduction of molecular genotyping methods in tuberculosis (TB) research in the early 1990s, it became possible to differentiate strain genotypes of M. tuberculosis infections (1–5). Molecular epidemiological studies showed that heterogenous, complex infections within a single individual can arise due to mixed infections with different strains of M. tuberculosis that are simultaneously or sequentially acquired (6, 7) or due to heterogenous infections where spontaneous mutations result in microevolution within the host (Fig. 1) (8–10). However, the classical molecular genotyping methods—spoligotyping (11), IS6110 restriction fragment length polymorphisms (RFLP) typing (12), and mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing (13, 14)—assessed this heterogeneity only by interrogating the number and distribution of repetitive elements in the M. tuberculosis genome. The use of Sanger sequencing (15) to target genes involved in drug resistance led to the discovery of heteroresistance (i.e., the simultaneous existence of drug-resistant and drug-sensitive M. tuberculosis subclones in the same sample) (16), but the detection of heterogeneity remained limited to that occurring in a small fraction of the M. tuberculosis genome. With the introduction of whole-genome sequencing (WGS), it became possible to study the entire genome, allowing comprehensive analyses of evolutionary relations.
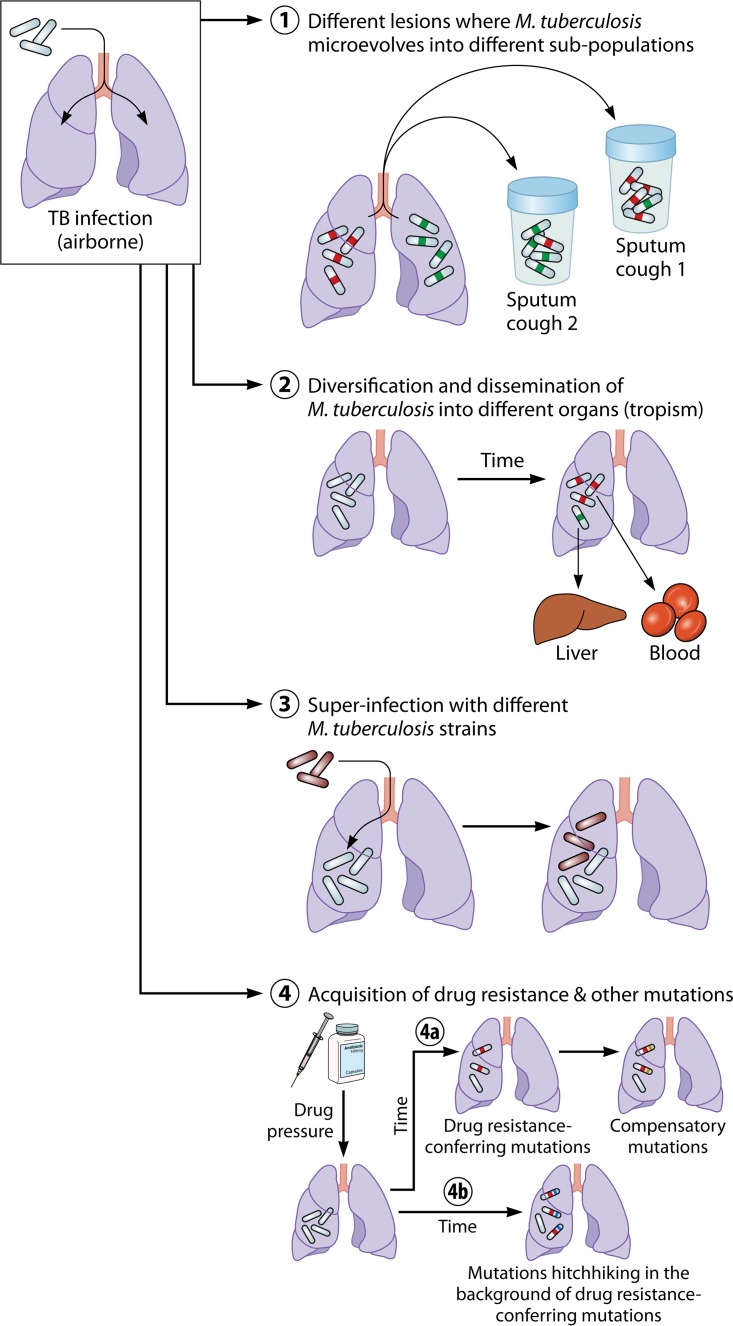
FIG 1.
Trajectory of within-host microevolution of M. tuberculosis. Four scenarios of microevolution of M. tuberculosis within a host are presented as follows. (Scenario 1) M. tuberculosis cells present in different lesions in the lung microevolve into distinct subpopulations through the acquisition of different single nucleotide variants (SNVs) (green and red). The different lesions might contribute bacilli to the sputum in different proportions, leading to the detection of different variants from distinct subpopulations. (Scenario 2) M. tuberculosis strains evolve in the lung and are subsequently disseminated into different organs, where they further evolve. Samples from different anatomical sites therefore harbor distinct M. tuberculosis subpopulations. (Scenario 3) An infected host is superinfected with a different but very similar M. tuberculosis strain, leading to the detection of different M. tuberculosis populations in subsequent samples. As the different M. tuberculosis strains are phylogenetically very similar, it is difficult to distinguish between within-host microevolution and exogenous reinfection with a new M. tuberculosis strain. (Scenario 4) Drug pressure leads to the acquisition of drug resistance-conferring mutations (red). (Scenario 4a) These mutations can lead to a loss of fitness which is subsequently restored through the acquisition of compensatory mutations (yellow). (Scenario 4b) Additional mutations might emerge in concert with the drug resistance-conferring mutations through genetic linkage to the latter (i.e., hitchhiking SNVs) (blue).
One of the biggest technical challenges is that of distinguishing between mixed infections with multiple similar strains and microevolution of strains resulting in minor clonal variation. Use of WGS on serial isolates to establish the chronology of mutation acquisition during an infection could help in distinguishing between these two types of within-host M. tuberculosis diversity and improve our understanding of M. tuberculosis infection dynamics. Misinterpreting microevolution can hamper TB control efforts, as transmission chains may be incorrectly determined and the driving forces of the TB epidemic might be misunderstood. Furthermore, the failure to detect heteroresistance impacts on the accuracy of phenotypic drug susceptibility testing and therefore also on treatment success. To date, there are limited data on the clinical relevance of microevolution within patients, especially pertaining to non-drug resistance-conferring mutations.
The aim of this review is to critically appraise the insights gained through WGS of serial and parallel (i.e., contemporarily collected) clinical M. tuberculosis isolates from the same patient, to ascertain the technical limitations, and to identify the remaining knowledge gaps about within-host microevolution in order to outline the way forward in this rapidly evolving field in tuberculosis research.
METHODS
Database Search, Selection of Eligible Articles, and Data Extraction
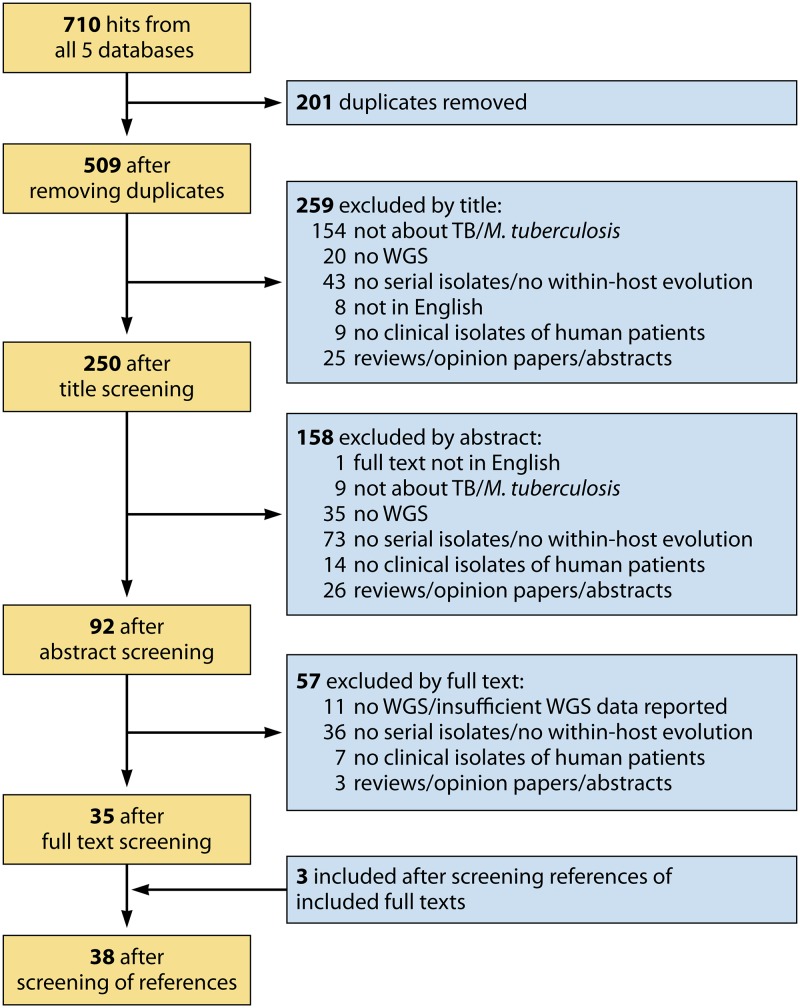
On 7 August 2018, the electronic databases PubMed, Web of Science Core Collection, Scopus, MEDLINE, and CINAHL were comprehensively searched using the terms: “tuberculosis,” “whole genome sequencing,” “within-host,” “serial,” and “parallel.” The detailed search strategy and the number of hits for each database are listed in Table S1 in the supplemental material. After removal of duplicates, the remaining titles, abstracts, and full manuscripts were independently screened by two authors. Publications were eligible for inclusion if they contained whole-genome sequencing data from serial or parallel M. tuberculosis isolates. Serial isolates were defined as clinical isolates from the same patient collected at different time points during the same TB episode or from initial and relapse episodes. Serial isolates from different infections (i.e., reinfection with a different M. tuberculosis strain) were excluded, as within-host microevolution is difficult to study in such isolates. Parallel isolates were defined as clinical isolates from the same patient collected at a single time point, e.g., several sputum samples or several samples collected from different anatomical sites in the body (e.g., sputum and blood samples). Where the decisions to include or exclude a specific publication differed between the two reviewers, the publication was discussed until a consensus was reached. In a second step, the reference lists of the included publications where screened for potentially missed articles. Non-English articles, reviews, opinion papers, abstracts, book chapters, studies focusing on mycobacteria other than M. tuberculosis or Mycobacterium africanum, studies conducted in animals, and studies analyzing in vitro isolates only (e.g., single colonies produced in culture) were excluded from this review (Fig. 2).
FIG 2.
PRISMA flow chart. A PRISMA flow chart describing the process for the selection of publications included in this review is presented. TB, tuberculosis; WGS, whole-genome sequencing.
Data extracted from each article included study topic, study site, study design, WGS methods (sequencing platform, sequencing depth, minimum coverage), other molecular methods used, analysis software and analysis parameters (reference strain, heterofrequency cutoff values, regions excluded from analysis, type of variants reported), mutation rate, loci affected by microevolution (gene, gene function, single nucleotide variant [SNV] position, amino acid change, frequency), genome stability and diversity, and information about the serial isolates (e.g., type of isolate, number of isolates per patient, lapse between times of collection of isolates, genotypic and phenotypic drug resistance profiles, drug pressure, SNV distance between serial isolates, and M. tuberculosis strain genotype).
Comparison of Microevolved Loci between Studies
A list of the genes and intergenic regions (IGRs) in which variants either emerged de novo or disappeared completely over time (i.e., the frequency of that variant was zero in at least one isolate of the patient) was compiled. Studies not reporting all variants detected or not providing sufficient information to assign an SNV to a specific patient and variants present in all serial isolates of a patient (even if changing in frequency over time) were excluded for this part of the analysis (Table S2). We then identified those genes and IGRs reported in two or more studies, as these genes and IGRs could have evolved convergently and might thus play a role in drug resistance, virulence, or compensatory mechanisms in M. tuberculosis. Variants in genes and IGRs reported more than twice were analyzed with the software Protein Variation Effect Analyzer (PROVEAN) (17), and literature and the mycobrowser database (18) were consulted to gain additional insight into their functionality and the protein’s mechanism of action.
RESULTS
Summary of Study Characteristics
The comprehensive search in five electronic databases resulted in 38 publications fulfilling all inclusion criteria (Table 1). The 38 publications reported on studies from 28 countries in five continents and covered diverse topics such as within-host evolution of drug resistance and compensatory mutations, M. tuberculosis genome stability and selection pressure, transmission dynamics, relapse versus reinfection, and diversity and heterogeneity of strains (Table 1). The sample sizes ranged from 2 to 9 serial and 2 to 140 parallel isolates collected from 1 to 178 patients, and the time between isolates spanned a few hours to several years (see Data Set S1 in the supplemental material). SNV distances ranged from 0 to 47 in studies that used a heterofrequency cutoff value of ≤30% and from 0 to 16 in studies using a heterofrequency cutoff value of ≥70%. Drug resistance profiles ranged from fully susceptible to extensively drug resistant (XDR; i.e., resistance against the two most effective first-line drugs, isoniazid [INH] and rifampin [RIF], plus resistance against one of the second-line-treatment fluoroquinolones and an injectable drug), with some isolates acquiring resistance during treatment. The M. tuberculosis strains belonged to five of the seven global lineages (lineages 1, 2, 3, 4, and 5), with strains of lineage 2 and lineage 4 being most prevalent. Six studies did not report the genotype of the analyzed M. tuberculosis strains.
TABLE 1.
Included articles
| Article | Date | Publication | Geographical area(s) of sample collection (sample origin) |
Main topic(s) |
|---|---|---|---|---|
| 1 | January 2011 | Saunders et al., Deep resequencing of serial sputum isolates of Mycobacterium tuberculosis during therapeutic failure due to poor compliance reveals stepwise mutation of key resistance genes on an otherwise stable genetic background (20) | United Kingdom | Drug resistance, genome stability |
| 2 | July 2012 | Comas et al., Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes (47) | Global (panels 1 + 2) Georgia, Uzbekistan, Kazakhstan (panel 3) |
Compensatory mutations |
| 3 | November 2012 | Walker et al., Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study (35) | United Kingdom | Outbreak, transmission |
| 4 | December 2012 | Sun et al., Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients (29) | China | Drug resistance and diversity |
| 5 | August 2013 | Farhat et al., Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis (25) | Italy | Drug resistance, compensatory mutations, selection pressure |
| 6 | November 2013 | Bryant et al., Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study (36) | Malaysia, South Africa, Thailand | Relapse versus reinfection |
| 7 | December 2013 | Clark et al., Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing (21) | Uganda | Relapse versus reinfection |
| 8 | December 2013 | Merker et al., Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients (23) | Germany, Georgia, Uzbekistan | Drug resistance |
| 9 | January 2014 | Pérez-Lago et al., Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission (40) | Spain | Transmission |
| 10 | October 2014 | Eldholm et al., Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient (22) | Argentina | Drug resistance |
| 11 | October 2014 | Guerra-Assunção et al., Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up (41) | Malawi | Relapse versus reinfection |
| 12 | February 2015 | Witney et al., Clinical application of whole-genome sequencing to inform treatment for multidrug-resistant tuberculosis cases (51) | United Kingdom | Drug resistance |
| 13 | March 2015 | Guerra-Assunção et al., Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area (37) | Malawi | Transmission |
| 14 | October 2015 | Black et al., Whole genome sequencing reveals genomic heterogeneity and antibiotic purification in Mycobacterium tuberculosis isolates (30) | South Africa | Heterogeneity |
| 15 | October 2015 | O’Neill et al., Diversity of Mycobacterium tuberculosis across evolutionary scales (39) | Argentina, Germany, Uzbekistan, Georgia, China | Diversity |
| 16 | November 2015 | Stinear et al., Genome sequence comparisons of serial multidrug-resistant Mycobacterium tuberculosis isolates over 21 years of infection in a single patient (38) | Australia | Drug resistance, genome stability |
| 17 | November 2015 | Pérez-Lago et al., Persistent infection by a Mycobacterium tuberculosis strain that was theorized to have advantageous properties, as it was responsible for a massive outbreak (28) | Spain | Genome stability, heterogeneity |
| 18 | November 2015 | Bloemberg et al., Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis (27) | Switzerland (Tibet) | Drug resistance |
| 19 | December 2015 | Liu et al., Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment (31) | China | Heterogeneity, compartmentalization |
| 20 | March 2016 | Silva Feliciano et al., Evaluation of resistance acquisition during tuberculosis treatment using whole genome sequencing (90) | Brazil | Drug resistance |
| 21 | June 2016 | Korhonen et al., Whole genome analysis of Mycobacterium tuberculosis isolates from recurrent episodes of tuberculosis, Finland, 1995-2013 (42) | Finland | Relapse versus reinfection |
| 22 | August 2016 | Faksri et al., Whole-genome sequencing analysis of serially isolated multi-drug and extensively drug resistant Mycobacterium tuberculosis from Thai patients (52) | Thailand | Drug resistance, persistence versus reinfection |
| 23 | August 2016 | Ssengooba et al., Whole genome sequencing reveals mycobacterial microevolution among concurrent isolates from sputum and blood in HIV infected TB patients (32) | Uganda | Heterogeneity, compartmentalization |
| 24 | August 2016 | Zhang et al., Genomic analysis of the evolution of fluoroquinolone resistance in Mycobacterium tuberculosis prior to tuberculosis diagnosis (91) | Taiwan | Drug resistance |
| 25 | October 2016 | Casali et al., Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: a retrospective observational study (43) | United Kingdom | Transmission |
| 26 | November 2016 | Lieberman et al., Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis (33) | South Africa | Heterogeneity, compartmentalization |
| 27 | October 2016 | Wollenberg et al., Whole genome sequencing of Mycobacterium tuberculosis provides insight into the evolution and genetic composition of drug-resistant tuberculosis in Belarus (92) | Belarus | Drug resistance |
| 28 | December 2016 | Datta et al., Longitudinal whole genome analysis of pre and post drug treatment Mycobacterium tuberculosis isolates reveals progressive steps to drug resistance (53) | Costa Rica, Spain, USA | Drug resistance |
| 29 | January 2017 | Manson et al., Mycobacterium tuberculosis whole genome sequences from southern India suggest novel resistance mechanisms and the need for region-specific diagnostics (46) | India | Drug resistance |
| 30 | January 2017 | Dheda et al., Outcomes, infectiousness, and transmission dynamics of patients with extensively drug-resistant tuberculosis and home-discharged patients with programmatically incurable tuberculosis: a prospective cohort study (93) | South Africa | Transmission |
| 31 | March 2017 | Witney et al., Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial (44) | South Africa, Zimbabwe, Botswana, Zambia | Relapse versus reinfection |
| 32 | April 2017 | Trauner et al., The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy (34) | China | Drug resistance, population dynamics |
| 33 | April 2017 | Navarro et al., In-depth characterization and functional analysis of clonal variants in a Mycobacterium tuberculosis strain prone to microevolution (19) | Spain | Direction of microevolution (heterogeneity) |
| 34 | July 2017 | Nsofor et al., Transmission is a noticeable cause of resistance among treated tuberculosis patients in Shanghai, China (26) | China | Drug resistance |
| 35 | August 2017 | Senghore et al., Whole-genome sequencing illuminates the evolution and spread of multidrug-resistant tuberculosis in Southwest Nigeria (94) | Nigeria | Drug resistance |
| 36 | November 2017 | Leung et al., Comparative genomic analysis of two clonally related multidrug resistant Mycobacterium tuberculosis by single molecule real time sequencing (48) | China | Drug resistance, growth fitness |
| 37 | January 2018 | Herranz et al., Mycobacterium tuberculosis acquires limited genetic diversity in prolonged infections, reactivations and transmissions Involving multiple hosts (45) | Spain and Latvia | Diversity |
| 38 | June 2018 | Dheda et al., Drug penetration gradients associated with acquired drug resistance in tuberculosis patients (24) | South Africa | Drug resistance |
Whole-Genome Sequencing Methods and Analysis Parameters
Most (24/38; 63%) studies complemented WGS data with (a combination of) spoligotyping, MIRU-VNTR typing, RFLP typing, targeted Sanger sequencing, or line probe assay data. WGS platforms, minimum required coverage levels, heterofrequency cutoff values, and additional filtering varied substantially between studies (see Table S3 in the supplemental material). Almost all studies (35/38; 89%) used the Illumina WGS platform, including HiSeq (20/38), MiSeq (6/38), Solexa (1/38), Genome Analyzer (1/38), MiSeq and HiSeq (3/38), HiSeq and Genome Analyzer (1/38), or an unspecified Illumina platform (3/38). The remaining three studies used the Ion Torrent platform, SOLiD 3, and SMRT (Single Molecule, Real-Time) sequencing of PacBio. Sequences were mostly frequently aligned using the Burrows Wheeler Aligner (BWA) (19/38; 50%), and more than half of the studies (21/38; 55%) used SAMtools for variant calling. The minimum required coverage of a loci ranged from 2× to 50× (Table S3); 10 studies did not report minimum coverage. The heterofrequency cutoff values applied ranged from no cutoff being applied to a cutoff value of 100% (Table S3); 14 studies did not report their heterofrequency cutoff. Most studies (82%) did not report on or systematically excluded repetitive regions (e.g., PE/PPE gene family) and transposable elements from their analysis.
Within-Host Evolution of Drug Resistance
All but one study (19) reported drug resistance levels, which ranged from fully susceptible to XDR (Data Set S1). Studies reported stepwise acquisition of resistance to INH and RIF (i.e., multidrug resistance [MDR]) over a period of 12 to 13 months (20, 21) and increases of resistance from para-aminosalicylic acid (PAS) monoresistance to XDR within 42 months (22) and from full susceptibility or MDR to XDR within 12 to 60 months (23, 24). Two other studies also reported drug resistance levels increasing from susceptible to MDR and from MDR to XDR, respectively, but without indicating the exact time lapse (25, 26). In the remaining 31 studies, no increase or a one-step increase in drug resistance level was observed during treatment. In the studies reporting details on the treatment regimen, drug resistance-conferring mutations were acquired during (partially) ineffective treatment (i.e., <4 effective drugs or treatment noncompliance). In a case study of a patient developing XDR-TB despite treatment compliance, retrospective WGS analysis revealed the stepwise acquisition of resistance-conferring mutations, including resistance to the new drugs bedaquiline and delamanid (27).
Diversity, Genome Stability, and Selection Pressure
Results on clonal variation and M. tuberculosis genome stability were conflicting. The first WGS study of serial M. tuberculosis isolates found no SNVs other than drug resistance-conferring mutations and concluded that M. tuberculosis genomes are highly stable within the host, with drug pressure hindering the diversification of the strains in loci other than the drug resistance-conferring ones (20). Similarly, another study showed that the M. tuberculosis genome can be very stable, remaining fully susceptible with no mutations emerging over 8 years, despite repeated treatment interruptions and treatment noncompliance over several years (28). In contrast, several other studies found higher levels of genetic divergence and M. tuberculosis was shown to have undergone a highly dynamic process of continuous evolution and purifying selection within a single host (22, 23, 27, 29–34). In one study, the SNVs between several coexisting subclones were all related to drug resistance (drug resistance-conferring mutations or fitness-restoring mutations) (23). In contrast, another study identified a range of coselected mutations hitchhiking in the background of emerging resistance-conferring mutations. In the latter study, the mutation rate was higher for drug resistance-conferring loci (7.0 SNVs/genome/year) than for the rest of the genome (1.1 SNVs/genome/year), demonstrating the increased selection pressure on drug resistance-conferring loci (22). The mutation rates were found to differ not only between different loci within the same M. tuberculosis strain but also between distinct strains, ranging from 0.3 to 7.0 SNVs per genome per year (22, 35–38). Sun et al. observed the appearance and disappearance of several clonal variants over time until the clone with the lowest fitness cost and the highest resistance level was fixed in the population (29). In two other studies, purifying selection was followed by rediversification either with (30) or without (33) drug pressure. Trauner et al. also observed high rates of genetic divergence and suggested that the stability of SNVs was not a function of their abundance but of drug pressure (34). Another study however, found an association between genetic diversification and disease severity but not with treatment or drug resistance phenotype (39). In that study, most loci showing extreme patterns of variation were found in drug resistance-conferring genes or in genes involved in regulation, synthesis, and transport of cell envelope lipids, suggesting a role of these genes in adaptation processes during infection and transmission (39). Other studies found spatial heterogeneity, with subclones of the same ancestor developing differently in different lesions or different areas of a lesion (24, 31), and also found repeated within-host dissemination of strains into different compartments (32, 33). Some of the detected minority clones were confined to specific areas of an organ (24, 33) and were unstable over time either with or without drug pressure (30, 36, 39).
SNV Distance between Serial Isolates To Infer Transmission and To Distinguish between Relapse and Reinfection
WGS of serial isolates from a patient has also been used to establish transmission chains and to distinguish relapses from exogenous reinfections (21, 35–37, 40–45). To infer direct transmission, the SNV distance between isolates of a transmission chain is determined. In the past, a maximum distance of <12 SNVs between any two strains was suggested to indicate genetic linkage and direct transmission (35). More-recent findings, however, have suggested that this threshold might not be valid in every setting and that several factors such as time lapse between isolates, selection pressure, mixed infections, or clonality of strains can impact on the number of SNVs emerging within a host, changing the distance between isolates (22, 33, 40, 43, 45). Furthermore, several studies found that the amount of within-host microevolution (i.e., SNV distance) can be as high as or higher than that seen between hosts along a transmission chain or in an outbreak (34, 37, 40, 43). Casali et al., for example, found that many strains along the transmission chain within an outbreak did not show any SNV difference despite a high level of within-host diversity and that transmission rarely resulted in the fixation of minor variants (43).
Five studies used WGS to distinguish relapse from exogenous reinfection (21, 36, 41, 42, 44). In those studies, relapse and reinfection could clearly be distinguished, with SNV distances of >100 to >1,400 for reinfections compared to an SNV distance of <9 SNVs for relapse cases (21, 36, 41, 42, 44). Only two cases showed intermediate SNV distances of 38 (42) and 57 (44) SNV differences, complicating the classification into relapse or reinfection. Following intensified WGS analyses, the latter could be classified as a relapse, as an initially unidentified minority subclone could be detected in the first isolate (44).
Emerging Variants and Their Function
Several studies investigated the function of specific emerging variants and found associations with drug resistance, compensatory mechanisms, virulence, or relapse and survival (Table 2). For example, variants in the fadD, fadE or pks gene families involved in cell wall biosynthesis pathways were repeatedly observed to evolve within the host (19, 22, 24, 29, 30, 32–34, 39, 42, 44, 46) and could play a role in fitness compensation or drug resistance (29, 39, 46). Emerging mutations in the bacA (Rv1819c) and Rv2326c genes, which code for putative ABC transporters, could be involved in resistance against aminoglycosides through the prolonged survival and reduced metabolic activity of M. tuberculosis (38). Investigating compensatory mechanisms, mutations in genes rpoA and rpoC were shown to appear only after the acquisition of a RIF resistance-conferring mutation in the rpoB gene (47). Another study showed that mutations in the genes Rv0888, Rv2071, and Rv3303c emerged in a highly resistant M. tuberculosis strain during treatment, potentially compensating for an initial decreased fitness of the bacteria (48).
TABLE 2.
Suggested functions of identified SNVs and their respective genes
| Suggested mechanism of action | Gene | SNV (amino acid change at codon) |
Study suggesting mechanism of action |
|---|---|---|---|
| Potential new drug resistance markers | ettA (Rv2477c) | W135G | Faksri et al. (52) |
| katG (Rv1908c) | L101R | Datta et al. (53) | |
| katG (Rv1908c) | A290P | Manson et al. (46) | |
| katG (Rv1908c) | L427P | Manson et al. (46) | |
| fadE24 (Rv3139) | R454S | Manson et al. (46) | |
| fabD (Rv2243) | A159T | Manson et al. (46) | |
| Associated with relapse and survival | eccB3 (Rv0283) | Witney et al. (44) | |
| mce1B (Rv0170) | Witney et al. (44) | ||
| bacA (Rv1819c) | Frame shift (insG) | Stinear et al. (38) | |
| Rv2326c | Frame shift (insC) | Stinear et al. (38) | |
| Potentially involved in compensatory mechanisms | rpoC (Rv0668) | Several | Comas et al. (47), Wollenberg et al. (92) |
| rpoA (Rv3457c) | Several | Comas et al. (47), Wollenberg et al. (92) | |
| Rv0888 | 360delG | Leung et al. (48) | |
| cobM (Rv2071) | 199_204del | Leung et al. (48) | |
| lpdA (Rv3303c) | V44I | Leung et al. (48) | |
| fadD32 (Rv3801c) | E444G | Sun et al. (29) | |
| fadE33 (Rv3564) | O'Neill et al. (39) | ||
| lprO (Rv0179c) | O'Neill et al. (39) | ||
| Associated with virulence | mce3R (Rv1963c) | Navarro et al. (19) | |
Comparison of Microevolved Loci between Studies
In the 22 studies presenting relevant and sufficient data (see Table S2 for the reasons for exclusion of 16 studies), we identified 1,101 different genes and IGRs in which variants either emerged de novo or disappeared over time in a patient (serial isolates) or existed in different anatomical sites of a patient (parallel isolates) (Data Set S2). Most (914; 83%) were reported from only a single study, 156 (14.2%) were reported from two studies, 21 (1.9%) from three studies, and 10 (0.9%) from 4 to 7 studies. The genes or IGRs identified in more than two studies included known compensatory and drug resistance-associated genes or IGRs (49, 50) such as rrs (MTB000019), rrl (MTB000020), gyrA (Rv0006), mshA (Rv0486), rpoB (Rv0667), rpoC (Rv0668), inhA promoter region (Rv1482c to Rv1484), fabG1/mabA (Rv1483), katG (Rv1908c), pncA (Rv2043c), ahpC (Rv2428), and embB (Rv3795) and 19 additional genes, many of which are associated with M. tuberculosis growth and the biosynthesis of cell wall lipids such as oxcA (Rv0118c) (29, 33, 34), PE_PGRS3 (Rv0278c) (24, 25, 33), Rv0457c (29, 34, 45), Rv0565c (23, 24, 29), Rv0726c (33, 34, 36), PE_PGRS9 (Rv0746) (24, 25, 51), prpR (Rv1129c) (33, 34, 36), pks4 (Rv1181) (32–34, 44), pks5 (Rv1527c) (19, 33, 34), fadD15 (Rv2187) (29, 33, 34), ettA (Rv2477) (42, 52, 53), Rv2024c (25, 29, 33), ppsA (Rv2931) (25, 33, 34), ppsE (Rv2935) (29, 30, 34, 47), recG (Rv2973c) (22, 34, 47), lpdA (Rv3303c) (34, 36, 48), glpK (Rv3696) (22, 30, 34, 42, 47, 52), espK (Rv3879c) (29, 33, 53), and mviN (Rv3910) (29, 33, 34, 42) (Table S4; see also Data Set S2). Mutations in those 19 non-drug resistance-associated genes were found in lineage 2 and lineage 4 strains and across different drug resistance levels. Most variants in these genes were detected in patients from whom only two serial or parallel isolates were available, thus preventing analysis of the fate of these mutations. A frequency increase across more than two serial isolates—patterns potentially resulting from within-host positive selection—was found only for a synonymous mutation in lpdA (34).
The potential functions of the identified genes differed. The lpdA and ettA genes may play a role in drug resistance. The missense mutation V44I in lpdA may destabilize the NAD(P)H quinone reductase LpdA (according to the DUET server but not the PROVEAN database) (18, 48). This destabilization would change the redox milieu, potentially interfering with the metabolization of the INH prodrug into its active form by the catalase-peroxidase KatG and resulting in an elevation of the INH MIC of the strain (48, 54). Trauner et al. detected two different lpdA variants present across several isolates from a patient infected with an XDR M. tuberculosis strain, only one of which emerged de novo. That variant remained at a very low frequency in all isolates and was accompanied by a lpdA promoter mutation with a frequency of 93% (34). Faksri et al. identified an emerging mutation in ettA (Rv2477c) that could represent a new marker for amikacin (AMI) and kanamycin (KAN) resistance (52) due to the encoded protein’s drug efflux pump activity and the involvement in macrolide antibiotics transport across membranes (18, 55–57). Two other studies observed that ettA mutations emerged in an INH-monoresistant strain (53) and in an INH-and-streptomycin-resistant strain (42), one of which appeared simultaneously with an INH resistance-conferring katG mutation (53).
Variants in the three genes ppsE, recG, and glpK appeared either to be nonbeneficial for the strain or to have evolved as a consequence of in vitro subculturing processing of the isolates rather than through within-host selection. Six studies identified variants in glpK (Rv3696), all observing the variants in one of the patient’s serial isolates only (22, 30, 34, 42, 47, 52) (Table S4). In addition, three contemporary isolates from the same patient carried several different unstable glpK mutations with heterofrequencies of ≤30%, removed from the population through purifying selection (34). The glycerol kinase GlpK is involved in glycerol metabolism and is associated with ESX-1 secretion (18, 58, 59). Bacteria without functioning GlpK cannot metabolize glycerol, which is not relevant during infection but is required in vitro with glycerol as a carbon source (58). PpsE is involved in the biosynthesis of phthiocerol dimycocerosate (PDIM), a virulence lipid on the outer cell membrane of M. tuberculosis, involved in phagocytosis and the prevention of phagosomal acidification (18, 60–62). Mutations in ppsE have been suggested to affect cell wall lipid synthesis and the strain’s mechanism to escape the host’s immune response. In vitro-passaged M. tuberculosis strain H37Rv was observed to become PDIM negative over time; in vivo, however, PDIM-negative M. tuberculosis strains are attenuated (63). The ATP-dependent DNA helicase RecG is induced in response to DNA-damaging agents (64) and plays a role in recombination and DNA repair (18, 64). M. tuberculosis strains carrying recG mutations may show erroneous replication and an increased mutation rate which can increase the chance of acquiring advantageous (e.g., drug resistance-conferring) or disadvantageous mutations.
The PE/PPE and PE-PGRS genes have been excluded in many of the studies; two members of the PE-PGRS gene family (PE-PGRS3 and PE-PGRS9) have nevertheless been detected in more than two studies (24, 25, 33, 51). PE-PGRS9 was found to be significantly induced in M. tuberculosis persisters in host cells and tissues (65). Farhat et al. detected four different variants in PE-PGRS9 appearing and disappearing over time (25), while the variant T320A emerged over the time of 9 months in another study (51). PE-PGRS3 has been found to be specifically expressed at low phosphate concentrations, and its unique, arginine-rich C-terminal domain was suggested to enhance adhesion and to be involved in tuberculosis pathogenesis (66).
Variants in the remaining 12 genes (oxcA, Rv0457c, Rv0565c, Rv0726c, prpR, pks4, pks5, Rv2024c, fadD15, ppsA, espK, and mviN) were detected in both serial and parallel isolates. The variants detected in parallel isolates were present in only few (<20%) isolates of a particular patient, with the exception of variants in the gene fadD15 (54/105; 51%). These variants showed highly variable frequencies ranging from 1% to 100%. In serial isolates, only variants in the genes ppsA, prpR, and Rv0565c were present in more than one isolate of a particular patient but these variants were transient and lost over time (23, 25, 34). PpsA, encoding a polyketide synthase, belongs to the same operon as ppsE and is also involved in the biosynthesis of PDIM (18, 67). PrpR is involved in fatty acid catabolism and replication initiation (68). Rv0565c is a putative monooxygenase. The R59H variant was suggested to be associated with prothionamide resistance due to their concurrent emergence (23). Sun et al. observed the emergence of the Rv0565c variant Y245C in an already ethionamide-resistant isolate (29), and the loss of Rv0565c did not lead to ethionamide resistance in an in vitro investigation (69). The genes pks4, pks5, and fadD15 are involved in the biosynthesis of fatty acids (pks4, pks5) and in their activation as acyl-coenzyme A (fadD15) (18). Mutations in genes of these gene families have been associated with compensatory mechanisms (Table 2) or INH resistance (pks5), based on the absence of such mutations in susceptible strains and association studies (70). mviN encodes a probable conserved transmembrane protein which has been found to be essential for in vitro growth (18). Variants in this gene detected in serial isolates are unstable and therefore appear not to be beneficial for the bacteria. The espK gene encodes a protein which has been associated with virulence (71), oxcA is involved in the catabolism of oxalic acid, Rv0726c encodes a possible S-adenosylmethionine-dependent methyltransferase, and the functions of Rv0457c and Rv2024c are still unknown (18).
DISCUSSION
WGS studies of serial and parallel M. tuberculosis isolates demonstrated that M. tuberculosis infections and their dynamics are much more intricate than previously thought. Various degrees of within-host microevolution and diversity were detected across studies, with some demonstrating the simultaneous presence of several transient subpopulations within the same host, whereas others found very stable M. tuberculosis genomes with no or only few emerging genomic changes over prolonged periods of treatment. The level of within-host microevolution can be as high as or higher than what is observed along transmission chains or in an outbreak. Drug resistance occurred stepwise in the presence of ineffective (<4 effective drugs) treatment. Most genes affected by emerging genetic changes were observed to carry variants in only a single study. The majority of genes repeatedly found to microevolve within the host across studies were either associated with drug resistance or involved in lipid synthesis, transport, or regulation, indicating a potential role in drug resistance mechanisms, virulence, and compensatory mechanisms.
Factors Influencing Variant Detection
Discrepancies in findings between studies can, at least in part, be explained by differences in factors influencing variant detection (Fig. 3). The number of and lapse between times of collection of serial isolates per patient within and between studies varied considerably (see Data Set S1 in the supplemental material) due to differences in the type and timing of sample collection, access to health care and treatment-seeking behavior of patients, treatment effectiveness, bacterial load, disease phenotype (e.g., degree of cavitation or pulmonary versus extrapulmonary TB), and time to culture conversion. A single sputum sample does not represent the whole diversity of an M. tuberculosis infection (31, 72), and culturing can lead to additional reductions of diversity (73, 74). Unstable minority clones detected only in a particular area of the lung—as observed by Lieberman et al.—would be missed, depending on the sampling time point, as would be subclones or distinct M. tuberculosis strains present only outside the lung (31, 33). If the lapse between the times of collection of two isolates is too long, the stepwise acquisition of mutations might be missed, not allowing the determination of the chronology and dynamics of events. If the lapse between the times of collection of two isolates is short, the observed diversity may reflect concurrently existing subclones rather than newly emerged mutations. If multiple samples are taken at the same time, different lesions may be sampled to different degrees. Differences in M. tuberculosis lineage could also influence differences between studies, as mutation rates have been shown to differ between M. tuberculosis lineages, with lineage 2 strains of the Beijing genotype having a higher mutation rate than lineage 4 strains (75). However, both stable and highly variable strains were detected in both lineages, leaving the observed differences unexplained.
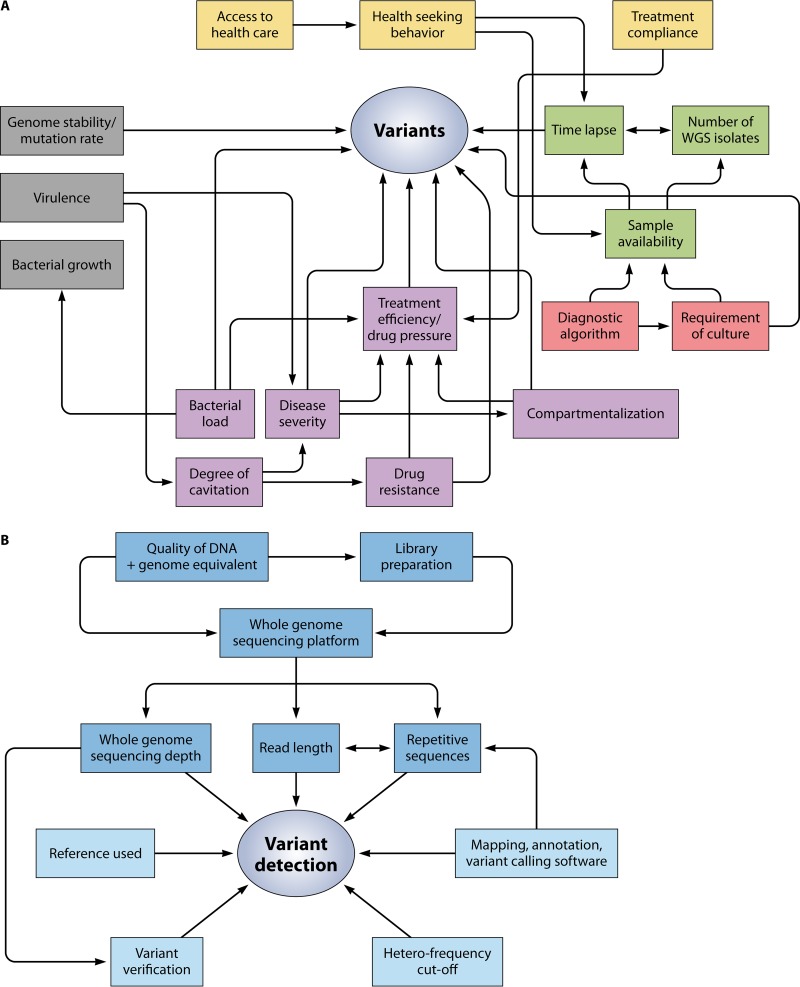
FIG 3.
Network of the various factors influencing the emergence and detection of variants. Several different factors influence the emergence of variants and their detection. (A) Bacterial factors (gray), patient aspects (yellow), sample characteristics (green), programmatic aspects (red), and clinical factors (purple) influencing variant detection and emergence. (B) Technical factors (dark blue) and analytical factors (light blue) influencing variant detection.
Differences in technical and analytical WGS approaches could further influence the differences in variant detection between studies. Sequencing depths and heterofrequency cutoff values differed between studies, but differences in the genomic stability of M. tuberculosis were observed even between studies with low heterofrequency cutoff values and deep sequencing (22, 28–30). The use of high heterofrequency cutoff values may result in missed low-frequency variants, while low heterofrequency cutoff values bear the risk of culture-induced variants or PCR and sequencing errors falsely being taken as representative of real variants. To reduce the risk of erroneous variant detection, Black et al. suggested a high confidence cutoff value of 30% for low-frequency variants based on a combined approach of WGS with an average sequencing depth of 137×, targeted Sanger sequencing, and statistical analysis (30). Increasing the sequencing depth would lead to more reads supporting a minor variant, decreasing the risk of detecting false positives and therefore allowing reduction of the heterofrequency cutoff value below the suggested threshold (76). Other strategies that can be used to confirm the validity of detected variants include Sanger sequencing and using more than one variant caller (i.e., inclusion of concurrently called variants only) (23, 29, 30). However, the former has been shown to be of limited utility for the detection of variants with a frequency below 30% (30, 77).
Lack of Standardized Reporting of WGS Results
The comparison of findings across studies and our ability to draw conclusions about common concepts of within-host microevolution and its impact on disease dynamics was also limited by the lack of standardized reporting of WGS analyses of serial isolates. Different filtering parameters, mappers, and variant callers with distinct mathematical algorithms were used across research groups, leading to differences in variant detection and interpretation of results. Furthermore, not all the studies included heterozygous loci, and many did not report the heterofrequency cutoff value, sequencing depth, accession number of the reference used, or all the SNVs (see Table S3 in the supplemental material). Synonymous SNVs were rarely reported, as these are considered to represent background mutations only. They can, however, affect cellular processes such as translation efficiency and internal promoters (78, 79). Hence, to improve our understanding of the role of synonymous SNVs in within-host microevolution, their detection should be reported. Furthermore, different databases of M. tuberculosis drug resistance markers exist and WGS analysis pipelines use different markers to determine the genetic drug susceptibility profile of an M. tuberculosis strain, leading to inconsistencies in the reporting of associations between SNVs and drug resistance. The use of a standardized pipeline, e.g., the pipeline developed by the Relational Sequencing TB Data Platform (80), as a reference to which alternative pipelines (developed specifically for studies investigating within-host microevolution) can be compared, the development of guidelines to report WGS results (similar to STROBE or CONSORT developed for reporting of observational or clinical trial studies), and an internationally standardized and curated list of (statistically) validated variants and their function would make findings more comparable.
Knowledge Gaps
While the WGS studies of serial and parallel isolates have explored several important topics and have contributed to an improved understanding of within-host microevolution of M. tuberculosis, many knowledge gaps remain (Table 3).
TABLE 3.
Identified knowledge gaps and recommendations for the way forward
| Limitations and knowledge gaps | Recommendations |
|---|---|
| Reporting of methods and results is often incomplete and not uniform across studies, and no standardized guidelines on reporting WGS results on serial and parallel isolates exist. This makes results irreproducible and difficult to compare, therefore not allowing to draw conclusions valid across studies. | Reporting of methods and results needs to be consistent. We recommend a minimum set of parameters to be reported by each study analyzing WGS of serial/parallel M. tuberculosis isolates. Technical parameters to be reported: • Culture process and library preparation (error risk assessment) • Sequencing platform used • Read length, coverage (whole genome), read depth • Paired or single reads • Mapping, annotation, and variant calling software used • Minimum coverage for minor variant and heterofrequency cutoff • Loci excluded from analysis • Additional filtering applied • Reference genome, including accession number Reporting of results should include: • Number, type, and location of SNVs between serial/parallel isolates • Heterofrequency of each SNV • All types of SNVs detected, including synonymous SNVs • Lapse between isolates • SNVs should clearly be assigned to the respective isolate and patient |
| WGS data analysis pipelines are not standardized and vary across studies, impacting on variant detection (95). While a standardized analysis pipeline might be beneficial for patient management and phylogenetic analyses, investigations on within-host microevolution require less-stringent filtering and in-depth analyses of raw data. | A standardized pipeline designed for patient management could be used as a reference to which results from individual analysis pipelines could be compared. For individual pipelines, filtering should be kept at a minimum to allow the detection of low-frequency variants and to determine underlying evolutionary dynamics. Deep (>1,000×) sequencing would further increase the detection of low-frequency variants. Results gained through this approach would provide valuable information to further develop and update the standardized pipeline for diagnostic purposes and patient management. |
| Our understanding of the physiological role and the phenotypic impact of within-host emerging variants is still limited, and our knowledge of drug resistance markers and phylogenetic markers is still incomplete. No internationally standardized list of markers is used to determine a comprehensive genotypic drug resistance profile of a strain. This potentially leads to false associations of SNVs with drug resistance or fitness compensation. | More laboratory-based experiments such as point mutagenesis and fitness assays are required to validate and confirm in silico-generated results and to determine the association between phenotype and genotype. Drug MICs of serial isolates should be measured and reported to determine a potential impact of an SNV on the level of drug resistance. To limit false associations of within-host emerging SNVs and drug resistance, a curated database containing a standardized list of validated variants should be developed, based on internationally acquired information from clinical M. tuberculosis isolates. |
| In vitro manipulation of clinical M. tuberculosis isolates can lead to the acquisition of variants that do not reflect within-host evolution. This can lead to an overestimation of within-host evolution and might complicate the analysis of potential new drug resistance markers or compensatory mutations. | Sequencing directly from sputum should be explored more widely to avoid culture-induced mutations. Eliminating the necessity to culture M. tuberculosis isolates prior to sequencing would furthermore decrease the time to results, an invaluable advantage in considering WGS in diagnostics in the future. |
| Only a few compensatory mechanisms are currently known. In particular, mechanisms compensating for loss of fitness in highly resistant M. tuberculosis strains are lacking. Studies analyzing serial isolates with increasing drug resistance across several resistance levels are limited, making it difficult to analyze the chronology of events during within-host emerging drug resistance. | Larger studies with serial isolates of different M. tuberculosis lineages and with increasing resistance across several levels are required to identify additional compensatory mechanisms. More-complex structural variants or epigenetic factors might be involved and should therefore be analyzed in the future. |
| Several factors such as drug pressure and disease severity have been suggested to drive M. tuberculosis within-host microevolution and diversity, but the contribution of each of these factors in different circumstances remains to be determined. | More data from longitudinal comparative WGS studies with serial M. tuberculosis isolates are required to study the impact of different (combinations of) bacterial, host, and clinical factors on within-host microevolution and diversity. |
| Exclusion of repetitive regions and large indels is common in Illumina WGS studies and leads to loss of information on about 20% of the genome. Therefore, the role of SNVs in repetitive areas in within-host microevolution and diversity is not known. | More data on regions difficult to map are required. Local reassembly approaches and long-read SMRT sequencing could be applied to include these genomic areas in the analyses and to gain more insights into the role of these genomic areas in microevolution. Combining SMRT and Illumina sequencing could reduce both error rates and loss of information. Using de novo reassembly instead of mapping to a reference genome would furthermore allow detection of SNVs and structural elements present in clinical M. tuberculosis strains but not in the reference used. |
| The impact of within-host evolution on transmission and M. tuberculosis strain relatedness is still not fully understood. Details about non-drug resistance-conferring SNVs emerging within the host are rarely reported in transmission studies or from analyses of recurrent episodes. Such SNVs might, however, influence the success of a strain (i.e., transmission and persistence in a patient) and change the SNV distance between strains, influencing our understanding of relatedness. | Within-host microevolution and the concurrent diversity of M. tuberculosis need to be considered in transmission and outbreak investigations. The type and location of detected SNVs should be analyzed and reported. Deep sequencing would allow investigation of whether and which low-frequency variants are transmitted and in addition would improve our ability to infer relatedness of two strains more accurately (i.e., to distinguish between relapse and reinfection). |
Drivers of within-host microevolution and diversity. Genetic drift and purifying selection have both been described as potential drivers of within-host microevolution and genetic diversity of M. tuberculosis, but our understanding of the drivers remains limited. Trauner et al. observed that sufficient drug pressure led to purifying selection of low-frequency variants and suggested that the stability of SNVs was not a function of their abundance (34). The latter is in conflict with the finding that the evolutionary fate of low-frequency variants was linked to allele frequency at the time of infection with M. tuberculosis in mice and Mycobacterium bovis BCG in humans (81). M. tuberculosis diversification was also shown to be correlated with disease severity (rather than drug pressure) (39) or with compartmentalization, with the degree of cavitation being an expression of disease severity (31). Future studies should investigate the role of bacterial load and host factors which, apart from HIV coinfection and treatment compliance, have not yet been addressed.
Repetitive areas and their role in adaptive evolution. There is a general lack of information about the role of repetitive areas (e.g., PE/PPE genes or insertion sequences) and large indels in adaptive evolution and diversification, as these were excluded from most studies given that they are difficult to map and cannot be distinguished from sequencing errors when using Illumina platforms. The SMRT sequencing technology produces long reads that can span these repetitive areas. It uses de novo assembly which allows the detection of rearrangements and copy number variations and the determination of allelic combinations but is prone to high error rates (48, 82, 83). Future studies should explore these repetitive areas, possibly using a combination of short-read and long-read sequencing to avoid the trade-off between loss of information and increased error rate.
Physiological role and phenotypic impact of microevolved variants. Apart from the known drug resistance-conferring mutations, the role and the impact of within-host emerging variants are not yet well understood. First, variants in genes associated with lipid synthesis, transport, or regulation have repeatedly been observed to evolve within the host (22, 29, 30, 34, 39, 42, 46, 52), suggesting that they may be targets of positive selection. Their role in adaptive evolution of M. tuberculosis should be further investigated. Second, the hypothesis of a potential role of emerging variants in the genes lpdA and ettA in drug resistance and compensation for loss of fitness was mainly based on the chronology of events and mostly not confirmed by MIC measurements for specific clones, fitness assays, or point mutagenesis experiments. Their role in resistance mechanisms and way of action should therefore be analyzed further.
Third, within-host emerging mutations have also been shown to hitchhike in the background of positively selected variants (22) or could represent culture artefacts (63). Our findings support this notion, as ppsE, recG, and glpK mutations were never found in more than one isolate of a patient—even in studies with more than two serial isolates—suggesting that these mutations may not be positively selected but may emerge as a result of subculturing of clinical isolates. WGS directly from sputum samples may avoid culture-induced artifacts, but few studies have succeeded in doing so (73, 84). More effort should be applied to improving the workflow from sample collection to sequencing results to obtain results representing the original state of the bacteria at the time of sample collection. Apart from circumventing the creation of false-positive variants, time to diagnosis would significantly be reduced (84).
Finally, the role of variants in multiple parallel isolates of a patient, i.e., in spatial heterogeneity, also requires further investigation. Such variants were detected in only a small proportion of sampled anatomical sites of a patient, in some at a very low frequency, in others fixed in the M. tuberculosis population. Specific variants might be beneficial in some anatomical sites but not in others and may impact the transmission of these variants, as M. tuberculosis strains located in extrapulmonary sites are not transmitted.
Compensatory mutations associated with drug resistance. While there are likely many compensatory mechanisms, maybe as many as there are drug resistance-conferring mutations, our understanding of compensatory mutations in M. tuberculosis remains limited to the rpoC/rpoA and ahpC promoter mutations (47, 85, 86). As MDR-TB and XDR-TB are mainly transmitted rather than acquired (26, 87, 88), large data sets from MDR strains evolving to pre-XDR and XDR within a patient are difficult to obtain. Yet studies of serial isolates would provide stronger evidence of causality than mere association studies, as the chronology of events can be studied, and compensatory mutations can be distinguished from hitchhiking ones. Large studies of (serial) isolates of patients infected with highly resistant M. tuberculosis strains of different lineages will be required to unravel the compensatory mechanisms in M. tuberculosis.
Impact of within-host microevolution on transmission. Within-host microevolution influences SNV distance between isolates, therefore complicating the interpretation of transmission directions and our ability to distinguish between relapse and reinfection. The latter may not always be possible, especially in high-burden countries with a clonal M. tuberculosis population structure, as despite a small SNV distance between two isolates, the possibility of reinfection with a very similar strain cannot be ruled out (89). Therefore, the SNV distance value used to infer genetic linkage might need to be context specific. Mixed infections with very similar strains—simultaneously or subsequently acquired—could also be a reason for intermediate SNV distances (>12 and <400 SNVs [35]) observed between serial isolates. Currently, the detected SNVs and evolving subclones cannot be assigned to a specific M. tuberculosis strain, and therefore the chronology and type of events cannot be established. This may potentially lead to misinterpretation of transmission chains or drug resistance profiles.
The impact of M. tuberculosis within-host microevolution in the context of transmission is generally not yet well understood. Within-host emerging SNVs might also play an important role in transmission dynamics, influencing the success of a strain. It remains largely unknown which factors make a strain transmissible, to what extent minor variants are transmitted, and whether transmissions follow a specific pattern (e.g., concerning M. tuberculosis strain genotype, frequency at time of infection, disease progression, etc.). To date, within-host microevolving SNVs have been used to distinguish between reinfection and relapse or between acquired and transmitted drug resistance or, numerically, to calculate the SNV distance and mutation rates in transmission studies, but their type, location, function, and frequency pattern are not systematically investigated. These knowledge gaps could be addressed by deep sequencing (>1,000×) of serial isolates of patients within a transmission cluster and comparing the identified variants, including low-frequency variants.
CONCLUSIONS
WGS studies of serial and parallel clinical M. tuberculosis isolates have demonstrated a wide range in diversity and stability of the M. tuberculosis genome within a single patient, but the mechanisms leading to this diversity and the function (other than drug resistance) of these emerging mutations remain poorly understood. Knowledge on the simultaneous presence and evolution of multiple M. tuberculosis subclones within a host and their clonal inference is, however, important as it can lead to heteroresistance and thus impact on patient management and surveillance and complicate public health efforts that rely on inferring transmission chains. In addition, it can interfere with the ability of clinical trials to determine the efficacy of new regimens, as such inferences require accurate distinction between treatment failure (relapse) and reinfection (transmission).
To improve our insights into the role of mutations in the successful adaptation of M. tuberculosis to the changing environment within a host, future studies should investigate the drivers of within-host genomic diversity; the role and function of emerging SNVs, including those in repetitive elements; the associations between different mutations; and their impact on phenotype, disease progression, diagnostics (e.g., MICs), and transmission. To enable comparisons between studies, technical approaches, WGS analyses, and reporting of serial and parallel M. tuberculosis isolates should be better standardized.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the South African Medical Research Council and the Swiss National Science Foundation (P2BSP3_165379) and by funding provided through the Flemish Fund for Scientific Research (FWO G0F8316N).
We declare that we have no conflict of interest.
Biographies

S. D. Ley received her B.Sc. and her M.Sc. in Molecular Biology from the University of Basel, Basel, Switzerland. For her Ph.D., she was mainly based at the Papua New Guinea Institute of Medical Research, Papua New Guinea, and completed her Ph.D. in Microbiology at the University of Basel. Until December 2018, Dr. Ley was as a postdoctoral fellow in the TB Genomics group of Professor Rob Warren, Division of Molecular Biology and Human Genetics at Stellenbosch University, Stellenbosch, South Africa. Her work focuses on the evolution and population structure of drug-resistant Mycobacterium tuberculosis within and between hosts using whole-genome sequencing. In January 2019, Dr. Ley joined the TB Research Unit at the Swiss Tropical and Public Health Institute, Basel, Switzerland, to continue her work on comparative genomics of highly resistant M. tuberculosis strains.

M. de Vos joined the Department of Biomedical Sciences at Stellenbosch University in 2007 to pursue a B.Sc. Honours degree, where she also completed her M.Sc. degree in 2009 and graduated with a Ph.D. in Molecular Biology in 2013. Her Ph.D. focused on the analysis of the whole genomes of closely related M. tuberculosis isolates to identify mechanisms regulating the intracellular concentration of rifampicin in M. tuberculosis. Margaretha is currently a postdoctoral fellow based at Stellenbosch University and a member of TORCH (Centre for Tuberculosis Omics Research). Her research focuses on the use of whole-genome sequencing to study the genomic evolution of M. tuberculosis during treatment and the acquisition of drug resistance with the aim to identify molecular markers and other risk factors for the prediction of treatment failure. Her other research interests include the development and validation of novel diagnostics for the identification of drug-resistant tuberculosis.

A. Van Rie completed her M.D. degree (1991) and Pediatrics Residency training (1996) at the University of Leuven in Belgium, after which she obtained her Ph.D. from the University of Stellenbosch in South Africa. From 2001 to 2015, she was a Professor of Epidemiology at the School of Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA. Since 2015, she has held an appointment as Professor of Epidemiology at the Department of Epidemiology and Social Medicine, Faculty of Medicine and Health Sciences, at the University of Antwerp in Belgium. For over 20 years, her work has focused on clinical, epidemiological, and translational tuberculosis research, with a special emphasis on TB/HIV, molecular epidemiology of drug-resistant TB, evaluation of new diagnostics, and implementation of research aimed at improving TB management and control in resource-poor high burden settings. Since 2017, Dr. Van Rie has been the Project Director of the Tuberculosis Omics Research (TORCH) Consortium.

R. M. Warren is currently the unit director of the South African Medical Research Council’s Centre for Tuberculosis Research. Under his guidance, the study of the molecular epidemiology of M. tuberculosis in a high-incidence setting (Cape Town, South Africa) was brought to the forefront of international tuberculosis research. Much of his work has provided new understanding which has allowed long-standing dogmas to be challenged. He has published more than 270 papers in international peer-reviewed journals in the fields of molecular epidemiology, drug resistance and bacterial evolution since 1996. His current research focuses on (i) the disease dynamics of drug-sensitive and M(X)DR-TB in the Western Cape, (ii) the development of novel diagnostics which are applicable to the developing world, (iii) discovery of the mechanisms whereby drug resistance develops, (iv) host-pathogen compatibility, (v) pathogen evolution, and (vi) mycobacterial epigenetics.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MMBR.00062-18.
REFERENCES
- 1.Horn DL, Hewlett D, Haas WH, Butler WR, Alfalla C, Tan E, Levine A, Nayak A, Opal SM. 1994. Superinfection with rifampin-isoniazid-streptomycin-ethambutol (RISE)-resistant tuberculosis in three patients with AIDS: confirmation by polymerase chain reaction fingerprinting. Ann Intern Med 121:115–116. doi: 10.7326/0003-4819-121-2-199407150-00007. [DOI] [PubMed] [Google Scholar]
- 2.Turett GS, Fazal BA, Justman JE, Alland D, Duncalf RM, Telzak EE. 1997. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis. Clin Infect Dis off Publ Infect Dis Soc Am 24:513–514. doi: 10.1093/clinids/24.3.513. [DOI] [PubMed] [Google Scholar]
- 3.Post FA, Willcox PA, Mathema B, Steyn LM, Shean K, Ramaswamy SV, Graviss EA, Shashkina E, Kreiswirth BN, Kaplan G. 2004. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J Infect Dis 190:99–106. doi: 10.1086/421501. [DOI] [PubMed] [Google Scholar]
- 4.Martín A, Herránz M, Serrano MJR, Bouza E, García de Viedma D. 2007. Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol 7:73. doi: 10.1186/1471-2180-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García de Viedma D, Marín M, Ruiz MJ, Bouza E. 2004. Analysis of clonal composition of Mycobacterium tuberculosis isolates in primary infections in children. J Clin Microbiol 42:3415–3418. doi: 10.1128/JCM.42.8.3415-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang R, Li X, Li J, Wu J, Shen X, Gui X, DeRiemer K, Liu L, Mei J, Gao Q. 2008. Mixed infections of Mycobacterium tuberculosis in tuberculosis patients in Shanghai, China. Tuberculosis (Edinb) 88:469–473. doi: 10.1016/j.tube.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rie A, Victor TC, Richardson M, Johnson R, van der Spuy GD, Murray EJ, Beyers N, Gey van Pittius NC, van Helden PD, Warren RM. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med 172:636–642. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Hajoj SAM, Akkerman O, Parwati I, Al-Gamdi S, Rahim Z, van Soolingen D, van Ingen J, Supply P, van der Zanden AGM. 2010. Microevolution of Mycobacterium tuberculosis in a tuberculosis patient. J Clin Microbiol 48:3813–3816. doi: 10.1128/JCM.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pérez-Lago L, Navarro Y, Herranz M, Bouza E, García-de-Viedma D. 2013. Differences in gene expression between clonal variants of Mycobacterium tuberculosis emerging as a result of microevolution. Int J Med Microbiol 303:674–677. doi: 10.1016/j.ijmm.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Navarro Y, Pérez-Lago L, Sislema F, Herranz M, de Juan L, Bouza E, García-de-Viedma D. 2013. Unmasking subtle differences in the infectivity of microevolved Mycobacterium tuberculosis variants coinfecting the same patient. Int J Med Microbiol 303:693–696. doi: 10.1016/j.ijmm.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol 36:762–771. [DOI] [PubMed] [Google Scholar]
- 14.Supply P, Magdalena J, Himpens S, Locht C. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol 26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Streicher EM, Bergval I, Dheda K, Böttger EC, Gey van Pittius NC, Bosman M, Coetzee G, Anthony RM, van Helden PD, Victor TC, Warren RM. 2012. Mycobacterium tuberculosis population structure determines the outcome of genetics-based second-line drug resistance testing. Antimicrob Agents Chemother 56:2420–2427. doi: 10.1128/AAC.05905-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi Y, Chan AP. 2015. PROVEAN Web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Navarro Y, Pérez-Lago L, Herranz M, Sierra O, Comas I, Sicilia J, Bouza E, García de Viedma D. 2017. In-depth characterization and functional analysis of clonal variants in a Mycobacterium tuberculosis strain prone to microevolution. Front Microbiol 8:694. doi: 10.3389/fmicb.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders NJ, Trivedi UH, Thomson ML, Doig C, Laurenson IF, Blaxter ML. 2011. Deep resequencing of serial sputum isolates of Mycobacterium tuberculosis during therapeutic failure due to poor compliance reveals stepwise mutation of key resistance genes on an otherwise stable genetic background. J Infect 62:212–217. doi: 10.1016/j.jinf.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Clark TG, Mallard K, Coll F, Preston M, Assefa S, Harris D, Ogwang S, Mumbowa F, Kirenga B, O’Sullivan DM, Okwera A, Eisenach KD, Joloba M, Bentley SD, Ellner JJ, Parkhill J, Jones-López EC, McNerney R. 2013. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS One 8:e83012. doi: 10.1371/journal.pone.0083012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eldholm V, Norheim G, von der Lippe B, Kinander W, Dahle UR, Caugant DA, Mannsåker T, Mengshoel AT, Dyrhol-Riise AM, Balloux F. 2014. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol 15:490. doi: 10.1186/s13059-014-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, Fattorini L, Oggioni MR, Cox H, Varaine F, Niemann S. 2013. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One 8:e82551. doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dheda K, Lenders L, Magombedze G, Srivastava S, Raj P, Arning E, Ashcraft P, Bottiglieri T, Wainwright H, Pennel T, Linegar A, Moodley L, Pooran A, Pasipanodya JG, Sirgel FA, van Helden PD, Wakeland E, Warren RM, Gumbo T. 1 November 2018. Drug penetration gradients associated with acquired drug resistance in tuberculosis patients. Am J Respir Crit Care Med doi: 10.1164/rccm.201711-2333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PKC, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nsofor CA, Jiang Q, Wu J, Gan M, Liu Q, Zuo T, Zhu G, Gao Q. 2017. Transmission is a noticeable cause of resistance among treated tuberculosis patients in Shanghai, China. Sci Rep 7:7691. doi: 10.1038/s41598-017-08061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloemberg GV, Keller PM, Stucki D, Stuckia D, Trauner A, Borrell S, Latshang T, Coscolla M, Rothe T, Hömke R, Ritter C, Feldmann J, Schulthess B, Gagneux S, Böttger EC. 2015. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 373:1986–1988. doi: 10.1056/NEJMc1505196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Lago L, Navarro Y, Montilla P, Comas I, Herranz M, Rodríguez-Gallego C, Ruiz Serrano MJ, Bouza E, García de Viedma D. 2015. Persistent infection by a Mycobacterium tuberculosis strain that was theorized to have advantageous properties, as it was responsible for a massive outbreak. J Clin Microbiol 53:3423–3429. doi: 10.1128/JCM.01405-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun G, Luo T, Yang C, Dong X, Li J, Zhu Y, Zheng H, Tian W, Wang S, Barry CE, Mei J, Gao Q. 2012. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis 206:1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black PA, de Vos M, Louw GE, van der Merwe RG, Dippenaar A, Streicher EM, Abdallah AM, Sampson SL, Victor TC, Dolby T, Simpson JA, van Helden PD, Warren RM, Pain A. 2015. Whole genome sequencing reveals genomic heterogeneity and antibiotic purification in Mycobacterium tuberculosis isolates. BMC Genomics 16:857. doi: 10.1186/s12864-015-2067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, Via LE, Luo T, Liang L, Liu X, Wu S, Shen Q, Wei W, Ruan X, Yuan X, Zhang G, Barry CE, Gao Q. 2015. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci Rep 5:17507. doi: 10.1038/srep17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ssengooba W, de Jong BC, Joloba ML, Cobelens FG, Meehan CJ. 5 August 2016. Whole genome sequencing reveals mycobacterial microevolution among concurrent isolates from sputum and blood in HIV infected TB patients. BMC Infect Dis doi: 10.1186/s12879-016-1737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman TD, Wilson D, Misra R, Xiong LL, Moodley P, Cohen T, Kishony R. 31 October 2016. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat Med doi: 10.1038/nm.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trauner A, Liu Q, Via LE, Liu X, Ruan X, Liang L, Shi H, Chen Y, Wang Z, Liang R, Zhang W, Wei W, Gao J, Sun G, Brites D, England K, Zhang G, Gagneux S, Barry CE, Gao Q. 2017. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol 18:71. doi: 10.1186/s13059-017-1196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TEA. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant JM, Harris SR, Parkhill J, Dawson R, Diacon AH, van Helden P, Pym A, Mahayiddin AA, Chuchottaworn C, Sanne IM, Louw C, Boeree MJ, Hoelscher M, McHugh TD, Bateson ALC, Hunt RD, Mwaigwisya S, Wright L, Gillespie SH, Bentley SD. 2013. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med 1:786–792. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerra-Assunção JA, Crampin AC, Houben RMGJ, Mzembe T, Mallard K, Coll F, Khan P, Banda L, Chiwaya A, Pereira RPA, McNerney R, Fine PEM, Parkhill J, Clark TG, Glynn JR. 2015. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife 4. doi: 10.7554/eLife.05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stinear TP, Denholm J, Globan M, Leslie D, Seemann T, Meumann EM, Porter JL, Fyfe JAM. 26 November 2015. Genome sequence comparisons of serial multi-drug-resistant Mycobacterium tuberculosis isolates over 21 years of infection in a single patient. Microb Genomics doi: 10.1099/mgen.0.000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Neill MB, Mortimer TD, Pepperell CS. 2015. Diversity of Mycobacterium tuberculosis across evolutionary scales. PLoS Pathog 11:e1005257. doi: 10.1371/journal.ppat.1005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pérez-Lago L, Comas I, Navarro Y, González-Candelas F, Herranz M, Bouza E, García-de-Viedma D. 2014. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J Infect Dis 209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 41.Guerra-Assunção JA, Houben RMGJ, Crampin AC, Mzembe T, Mallard K, Coll F, Khan P, Banda L, Chiwaya A, Pereira RPA, McNerney R, Harris D, Parkhill J, Clark TG, Glynn JR. 2015. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis 211:1154–1163. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korhonen V, Smit PW, Haanperä M, Casali N, Ruutu P, Vasankari T, Soini H. 2016. Whole genome analysis of Mycobacterium tuberculosis isolates from recurrent episodes of tuberculosis, Finland, 1995–2013. Clin Microbiol Infect 22:549–554. doi: 10.1016/j.cmi.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Casali N, Broda A, Harris SR, Parkhill J, Brown T, Drobniewski F. 2016. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: a retrospective observational study. PLoS Med 13:e1002137. doi: 10.1371/journal.pmed.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witney AA, Bateson ALE, Jindani A, Phillips PPJ, Coleman D, Stoker NG, Butcher PD, McHugh TD; RIFAQUIN Study Team. 2017. Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med 15:71. doi: 10.1186/s12916-017-0834-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herranz M, Pole I, Ozere I, Chiner-Oms Á, Martínez-Lirola M, Pérez-García F, Gijón P, Serrano MJR, Romero LC, Cuevas O, Comas I, Bouza E, Pérez-Lago L, García-de-Viedma D. 2017. Mycobacterium tuberculosis acquires limited genetic diversity in prolonged infections, reactivations and transmissions involving multiple hosts. Front Microbiol 8:2661. doi: 10.3389/fmicb.2017.02661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manson AL, Abeel T, Galagan JE, Sundaramurthi JC, Salazar A, Gehrmann T, Shanmugam SK, Palaniyandi K, Narayanan S, Swaminathan S, Earl AM. 2017. Mycobacterium tuberculosis whole genome sequences from southern India suggest novel resistance mechanisms and the need for region-specific diagnostics. Clin Infect Dis 64:1494–1501. doi: 10.1093/cid/cix169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2012. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung KS-S, Siu GK-H, Tam KK-G, To SW-C, Rajwani R, Ho P-L, Wong SS-Y, Zhao WW, Ma OC-K, Yam W-C. 2017. Comparative genomic analysis of two clonally related multidrug resistant Mycobacterium tuberculosis by single molecule real time sequencing. Front Cell Infect Microbiol 7:478. doi: 10.3389/fcimb.2017.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. 2014. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, Hanna D, Kim PS, Liwski R, Zignol M, Gilpin C, Niemann S, Denkinger CM, Fleming J, Warren RM, Crook D, Posey J, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Murray M, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, Ranganathan UDK, McNerney R, Ezewudo M, Cirillo DM, Schito M, Köser CU, Rodwell TC. 2017. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 50. doi: 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witney AA, Gould KA, Arnold A, Coleman D, Delgado R, Dhillon J, Pond MJ, Pope CF, Planche TD, Stoker NG, Cosgrove CA, Butcher PD, Harrison TS, Hinds J. 2015. Clinical application of whole-genome sequencing to inform treatment for multidrug-resistant tuberculosis cases. J Clin Microbiol 53:1473–1483. doi: 10.1128/JCM.02993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faksri K, Tan JH, Disratthakit A, Xia E, Prammananan T, Suriyaphol P, Khor CC, Teo Y-Y, Ong RT-H, Chaiprasert A. 2016. Whole-genome sequencing analysis of serially isolated multi-drug and extensively drug resistant Mycobacterium tuberculosis from Thai patients. PLoS One 11:e0160992. doi: 10.1371/journal.pone.0160992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Datta G, Nieto LM, Davidson RM, Mehaffy C, Pederson C, Dobos KM, Strong M. 2016. Longitudinal whole genome analysis of pre and post drug treatment Mycobacterium tuberculosis isolates reveals progressive steps to drug resistance. Tuberculosis (Edinb) 98:50–55. doi: 10.1016/j.tube.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Vilchèze C, Weisbrod TR, Chen B, Kremer L, Hazbón MH, Wang F, Alland D, Sacchettini JC, Jacobs WR. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother 49:708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel J, Abraham L, Martin A, Pablo X, Reyes S. 31 October 2017. Rv2477c is an antibiotic-sensitive manganese-dependent ABC-F ATPase in Mycobacterium tuberculosis. Biochem Biophys Res Commun doi: 10.1016/j.bbrc.2017. [DOI] [PubMed] [Google Scholar]
- 56.Kerr ID. 2004. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun 315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 57.Gupta AK, Katoch VM, Chauhan DS, Sharma R, Singh M, Venkatesan K, Sharma VD. 2010. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb Drug Resist Larchmt N 16:21–28. doi: 10.1089/mdr.2009.0054. [DOI] [PubMed] [Google Scholar]
- 58.Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, Jiricek J, Keller TH, Chatterjee A, Tan MP, Ujjini M, Rao SPS, Camacho L, Bifani P, Mak PA, Ma I, Barnes SW, Chen Z, Plouffe D, Thayalan P, Ng SH, Au M, Lee BH, Tan BH, Ravindran S, Nanjundappa M, Lin X, Goh A, Lakshminarayana SB, Shoen C, Cynamon M, Kreiswirth B, Dartois V, Peters EC, Glynne R, Brenner S, Dick T. 2010. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Comms 1:1. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Astarie-Dequeker C, Le Guyader L, Malaga W, Seaphanh F-K, Chalut C, Lopez A, Guilhot C. 2009. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog 5:e1000289. doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao A, Ranganathan A. 2004. Interaction studies on proteins encoded by the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Mol Genet Genomics 272:571–579. doi: 10.1007/s00438-004-1088-3. [DOI] [PubMed] [Google Scholar]
- 62.Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. 2005. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell 17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Domenech P, Reed MB. 2009. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiol Read Engl 155:3532–3543. doi: 10.1099/mic.0.029199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thakur RS, Basavaraju S, Somyajit K, Jain A, Subramanya S, Muniyappa K, Nagaraju G. 2013. Evidence for the role of Mycobacterium tuberculosis RecG helicase in DNA repair and recombination. FEBS J 280:1841–1860. doi: 10.1111/febs.12208. [DOI] [PubMed] [Google Scholar]
- 65.Delogu G, Sanguinetti M, Pusceddu C, Bua A, Brennan MJ, Zanetti S, Fadda G. 2006. PE_PGRS proteins are differentially expressed by Mycobacterium tuberculosis in host tissues. Microbes Infect 8:2061–2067. doi: 10.1016/j.micinf.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 66.De Maio F, Battah B, Palmieri V, Petrone L, Corrente F, Salustri A, Palucci I, Bellesi S, Papi M, Rubino S, Sali M, Goletti D, Sanguinetti M, Manganelli R, De Spirito M, Delogu G. 2018. PE_PGRS3 of Mycobacterium tuberculosis is specifically expressed at low phosphate concentration, and its arginine-rich C-terminal domain mediates adhesion and persistence in host tissues when expressed in Mycobacterium smegmatis. Cell Microbiol 20:e12952. doi: 10.1111/cmi.12952. [DOI] [PubMed] [Google Scholar]
- 67.Bisson GP, Mehaffy C, Broeckling C, Prenni J, Rifat D, Lun DS, Burgos M, Weissman D, Karakousis PC, Dobos K. 2012. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis. J Bacteriol 194:6441–6452. doi: 10.1128/JB.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masiewicz P, Brzostek A, Wolański M, Dziadek J, Zakrzewska-Czerwińska J. 2012. A novel role of the PrpR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS One 7:e43651. doi: 10.1371/journal.pone.0043651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grant SS, Wellington S, Kawate T, Desjardins CA, Silvis MR, Wivagg C, Thompson M, Gordon K, Kazyanskaya E, Nietupski R, Haseley N, Iwase N, Earl AM, Fitzgerald M, Hung DT. 2016. Baeyer-Villiger monooxygenases EthA and MymA are required for activation of replicating and non-replicating Mycobacterium tuberculosis inhibitors. Cell Chem Biol 23:666–677. doi: 10.1016/j.chembiol.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shekar S, Yeo ZX, Wong JCL, Chan MKL, Ong DCT, Tongyoo P, Wong S-Y, Lee ASG. 2014. Detecting novel genetic variants associated with isoniazid-resistant Mycobacterium tuberculosis. PLoS One 9:e102383. doi: 10.1371/journal.pone.0102383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. 2007. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog 3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shamputa IC, Jugheli L, Sadradze N, Willery E, Portaels F, Supply P, Rigouts L. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir Res 7:99. doi: 10.1186/1465-9921-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doyle RM, Burgess C, Williams R, Gorton R, Booth H, Brown J, Bryant JM, Chan J, Creer D, Holdstock J, Kunst H, Lozewicz S, Platt G, Romero EY, Speight G, Tiberi S, Abubakar I, Lipman M, McHugh TD, Breuer J. 26 July 2018. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J Clin Microbiol doi: 10.1128/JCM.00666-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Metcalfe JZ, Streicher E, Theron G, Colman RE, Penaloza R, Allender C, Lemmer D, Warren RM, Engelthaler DM. 2017. Mycobacterium tuberculosis subculture results in loss of potentially clinically relevant heteroresistance. Antimicrob Agents Chemother 61:e00888-17. doi: 10.1128/AAC.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, Flynn JL, Fortune SM. 2011. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. 2014. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 77.Feuerriegel S, Schleusener V, Beckert P, Kohl TA, Miotto P, Cirillo DM, Cabibbe AM, Niemann S, Fellenberg K. 2015. PhyResSE: a Web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol 53:1908–1914. doi: 10.1128/JCM.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kahali B, Basak S, Ghosh TC. 2007. Reinvestigating the codon and amino acid usage of S. cerevisiae genome: a new insight from protein secondary structure analysis. Biochem Biophys Res Commun 354:693–699. doi: 10.1016/j.bbrc.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 80.Ezewudo M, Borens A, Chiner-Oms Á, Miotto P, Chindelevitch L, Starks AM, Hanna D, Liwski R, Zignol M, Gilpin C, Niemann S, Kohl TA, Warren RM, Crook D, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, McNerney R, Cirillo DM, Schito M, Rodwell TC, Posey J. 2018. Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase. Sci Rep 8:15382. doi: 10.1038/s41598-018-33731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Copin R, Wang X, Louie E, Escuyer V, Coscolla M, Gagneux S, Palmer GH, Ernst JD. 2016. Within host evolution selects for a dominant genotype of Mycobacterium tuberculosis while T cells increase pathogen genetic diversity. PLoS Pathog 12:e1006111. doi: 10.1371/journal.ppat.1006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 83.Roberts RJ, Carneiro MO, Schatz MC. 2013. The advantages of SMRT sequencing. Genome Biol 14:405. doi: 10.1186/gb-2013-14-6-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Votintseva AA, Bradley P, Pankhurst L, del Ojo Elias C, Loose M, Nilgiriwala K, Chatterjee A, Smith EG, Sanderson N, Walker TM, Morgan MR, Wyllie DH, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2017. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sherman DR, Mdluli K, Hickey MJ, Arain TM, Morris SL, Barry CE, Stover CK. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 86.de Vos M, Müller B, Borrell S, Black PA, van Helden PD, Warren RM, Gagneux S, Victor TC. 2013. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother 57:827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shah NS, Auld SC, Brust JCM, Mathema B, Ismail N, Moodley P, Mlisana K, Allana S, Campbell A, Mthiyane T, Morris N, Mpangase P, van der Meulen H, Omar SV, Brown TS, Narechania A, Shaskina E, Kapwata T, Kreiswirth B, Gandhi NR. 2017. Transmission of extensively drug-resistant tuberculosis in South Africa. N Engl J Med 376:243–253. doi: 10.1056/NEJMoa1604544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang F, Shao L, Fan X, Shen Y, Diao N, Jin J, Sun F, Wu J, Chen J, Weng X, Cheng X, Zhang Y, Zhang W. 2015. Evolution and transmission patterns of extensively drug-resistant tuberculosis in China. Antimicrob Agents Chemother 59:818–825. doi: 10.1128/AAC.03504-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drobniewski F, Nikolayevskyy V, Maxeiner H, Balabanova Y, Casali N, Kontsevaya I, Ignatyeva O. 2013. Rapid diagnostics of tuberculosis and drug resistance in the industrialized world: clinical and public health benefits and barriers to implementation. BMC Med 11:190. doi: 10.1186/1741-7015-11-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Silva Feliciano C, Rodrigues Plaça J, Peronni K, Araújo Silva W, Roberto Bollela V. 2016. Evaluation of resistance acquisition during tuberculosis treatment using whole genome sequencing. Braz J Infect Dis 20:290–293. doi: 10.1016/j.bjid.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang D, Gomez JE, Chien J-Y, Haseley N, Desjardins CA, Earl AM, Hsueh P-R, Hung DT. 2016. Genomic analysis of the evolution of fluoroquinolone resistance in Mycobacterium tuberculosis prior to tuberculosis diagnosis. Antimicrob Agents Chemother 60:6600–6608. doi: 10.1128/AAC.00664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wollenberg KR, Desjardins CA, Zalutskaya A, Slodovnikova V, Oler AJ, Quiñones M, Abeel T, Chapman SB, Tartakovsky M, Gabrielian A, Hoffner S, Skrahin A, Birren BW, Rosenthal A, Skrahina A, Earl AM. 2016. Whole-genome sequencing of Mycobacterium tuberculosis provides insight into the evolution and genetic composition of drug-resistant tuberculosis in Belarus. J Clin Microbiol 55:e02116-16. doi: 10.1128/JCM.02116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dheda K, Limberis JD, Pietersen E, Phelan J, Esmail A, Lesosky M, Fennelly KP, Te Riele J, Mastrapa B, Streicher EM, Dolby T, Abdallah AM, Ben-Rached F, Simpson J, Smith L, Gumbo T, van Helden P, Sirgel FA, McNerney R, Theron G, Pain A, Clark TG, Warren RM. 19 January 2017. Outcomes, infectiousness, and transmission dynamics of patients with extensively drug-resistant tuberculosis and home-discharged patients with programmatically incurable tuberculosis: a prospective cohort study. Lancet Respir Med doi: 10.1016/S2213-2600(16)30433-7. [DOI] [PubMed] [Google Scholar]
- 94.Senghore M, Otu J, Witney A, Gehre F, Doughty EL, Kay GL, Butcher P, Salako K, Kehinde A, Onyejepu N, Idigbe E, Corrah T, de Jong B, Pallen MJ, Antonio M. 2017. Whole-genome sequencing illuminates the evolution and spread of multidrug-resistant tuberculosis in Southwest Nigeria. PLoS One 12:e0184510. doi: 10.1371/journal.pone.0184510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S. 2017. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci Rep 7:46327. doi: 10.1038/srep46327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.