Abstract
Biology arises from the crowded molecular environment of the cell, rendering it a challenge to understand biological pathways based on the reductionist, low-concentration in vitro conditions generally employed for mechanistic studies. Recent evidence suggests that low-affinity interactions between cellular biopolymers abound, with still poorly defined effects on the complex interaction networks that lead to the emergent properties and plasticity of life. The author uses mass-action considerations to underscore that the sheer number of weak interactions expected from the complex mixture of cellular components significantly shapes biological pathway specificity. In particular, on-pathway – i.e., “functional” – become those interactions thermodynamically and kinetically sufficiently stable to survive the incessant onslaught of the many off-pathway (“non-functional”) interactions. Consequently, to better understand the molecular biology of the cell a further paradigm shift is needed toward mechanistic experimental and computational approaches that probe intracellular diversity and complexity more directly.
Keywords: biological specificity, biomolecular machines, kinetic proofreading, law of mass action, network analysis, systems biology
Graphical Abstract
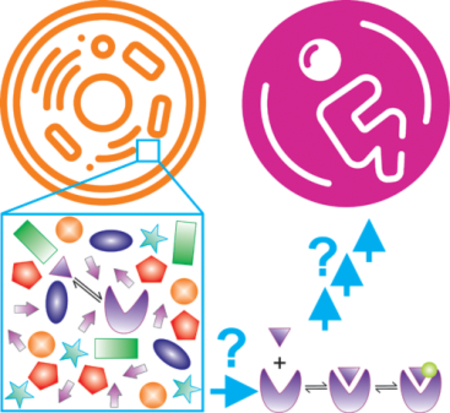
How exactly specific biological pathways and eventually life arise from the crowded molecular environment of the cell is a problem that will require a paradigm shift toward mechanistic experimental and computational approaches that probe intracellular diversity and complexity more directly.
1. Introduction
Linus Pauling – the famed chemist, biochemist, peace activist, author, educator, double Nobel laureate and co-founder of fields as diverse as quantum chemistry and molecular biology – noted that “biological specificity is the major problem about understanding life”,[1] a statement that applies both to organismal species and the specificity of molecular pathways in the cell. In the wake of the human genome project, advances in high-throughput transcriptome and proteome analysis are giving us rapidly expanding views of the sheer multitude of biopolymers found in a cell all at once, leading to major efforts to create comprehensive “cell atlas” catalogues.[2–8] These advances raise the critical problem: How does such a deluge of intracellular players lead to the emergence of specific molecular pathways from the “noise” of the billions of dynamic hetero-bimolecular interactions possible, most of which would seemingly not contribute to cell survival and reproduction?
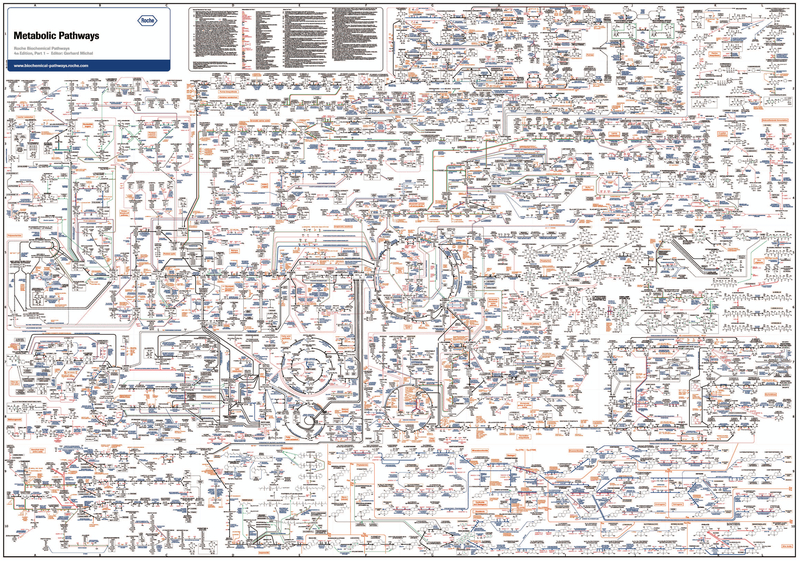
The human mind is bound to think in linear cause-and-effect paradigms since many events in our everyday experience appear to be the result of identifiable causes. Consequently, the basic assignment of functions to the many non-protein coding (or non-coding) RNAs discovered to be encoded in our genome, for example, is seen as tied to the unresolved question of whether or not they participate in any specific intracellular pathway, with the expectation that their removal should lead to identifiable phenotypic changes.[9,10] Similarly, bioscience textbooks teach that the specificity of, say, an enzyme is a direct consequence of its molecular structure in relation to only that of its specific substrate; and that a metabolic pathway is a very specific sequence of reactions linked through a linear flux of interrelated substrates. While breaking down biology into a linked series of simple cause-and-effect relationships facilitates human comprehension, a single look at a map of the main metabolic pathways in a cell reminds us that the cellular reality is more complex (Fig. 1).[11] This insight led to the rise of holistic systems biology approaches aiming to understand the complexity of biology based on the notion that cellular networks interact in synergistic ways that make the whole more than the sum of its parts. As Denis Noble, one of the recognized “godfathers” of systems biology, writes:[12] “This…is the challenge that sequencing the genome has raised. Can we put Humpty-Dumpty back together again? That is where ‘systems biology’ comes in [– a new approach that] is about putting together rather than taking apart, integration rather than reduction.”
Figure 1.
The generic biochemical pathway map of the cell. Reproduced with permission from F. Hoffmann-La Roche Ltd. For a detailed, interactive version of this map, please visit https://www.roche.com/sustainability/philanthropy/science_education/pathways.htm.
It is also increasingly appreciated that biology features a pervasive element of stochasticity, leading to diverse, spatiotemporally inhomogeneous distributions of molecules both within and across individual cells, even when members of a clonal cell line or tumor.[13–17] Yet, this growing appreciation of the non-linearity and stochasticity that drive and distinguish biological systems has so far arguably not influenced as much our understanding of what makes the cell “pick and choose,” or evolve, specific metabolic, signaling, or other pathways.
This essay aims to inspire further efforts to probe the physical basis of biological specificity of cellular pathways by integrating our classical knowledge from reductionist studies of the pairwise binding interactions of purified biopolymers in vitro with the emerging picture of the cell’s complex, crowded molecular environment. Simple considerations based on the law of mass action underscore that the plethora of diverse, weak (“non-specific”) interaction partners will have a significant impact on any specific intermolecular complex in the cell. Increasingly utilizing the cell itself as the reaction vessel of choice for mechanistic studies therefore promises to yield a refined biological specificity paradigm deeply dependent on cellular context and the full diversity of all molecular interactions, both strong and weak. Notably, rather than seeking to provide a comprehensive review of biology, the essay aims to connect ideas and provoke new thought.
2. The Reductionist Law of Mass Action, Expanded
When a natural law is first described in 1864,[18] forgotten, then rediscovered in 1877 by the famous Henry van’t Hoff,[19] and remains accepted over a century-and-a-half later, it has to be foundational. The law of mass action predicts the behavior of a chemical reaction in dynamic equilibrium, in that the ratio of the concentrations of products divided by those of the reactants is constant for a given temperature and pressure―
For a reaction of the stoichiometry: aA + bB ⇌ cC + dD
The equilibrium constant K is:
Importantly, Waage and Guldberg[18] in 1864 already recognized that a chemical equilibrium is dynamic such that the ratio of the rate constant of the forward reaction divided by that of the reverse reaction equals the equilibrium constant―[18]
This observation gives rise to Le Chatelier’s principle whereby a system at equilibrium that is subjected to a change in concentration, temperature or pressure readjusts to counteract the effect of the applied change until a new equilibrium is established. Strikingly, while not without exceptions at short time scales[20], Le Chatelier’s principle has found applications in fields as diverse as economy[21] and mechanical engineering (per Newton’s third law: “For every action, there is an equal and opposite reaction”) in that a system exposed to stress will generally respond by reducing that stress. Such dynamic adaptation is also one key to understanding biological specificity.
Suppose a cell contains two binding partners S and L of a complex (such as two proteins, a protein and an RNA or DNA, or two RNAs) at the typically low intracellular concentrations of picomolar (for DNA in a mammalian cell) to micromolar (for a highly expressed particle such as the ribosome), with their dissociation equilibrium constant Kd,S having evolved to be similar to the concentration of the limiting binding partner, say L. Then, the limiting component L will have a 50% probability to be in complex with its excess binding partner S at any given time. Suppose further that a second binding partner C of dissociation equilibrium constant Kd,C that coincidentally has the same value as Kd,S now becomes expressed to the same cellular concentration as S. It follows that through the ensuing two equilibria, which are coupled by sharing L, S and C are effectively competing for L such that each occupies L ~33% of the time―
S + L ⇌ SL Leads to an equilibrium dissociation constant of:
C + L ⇌ CL Leads to an equilibrium dissociation constant of:
Together, these coupled equilibria lead to the relation: , which in our example of Kd,S = Kd,C equals 1; and with [S] = [C] and [CL] + [SL] + [L] = [L]total follows [CL] = [SL] = [L].
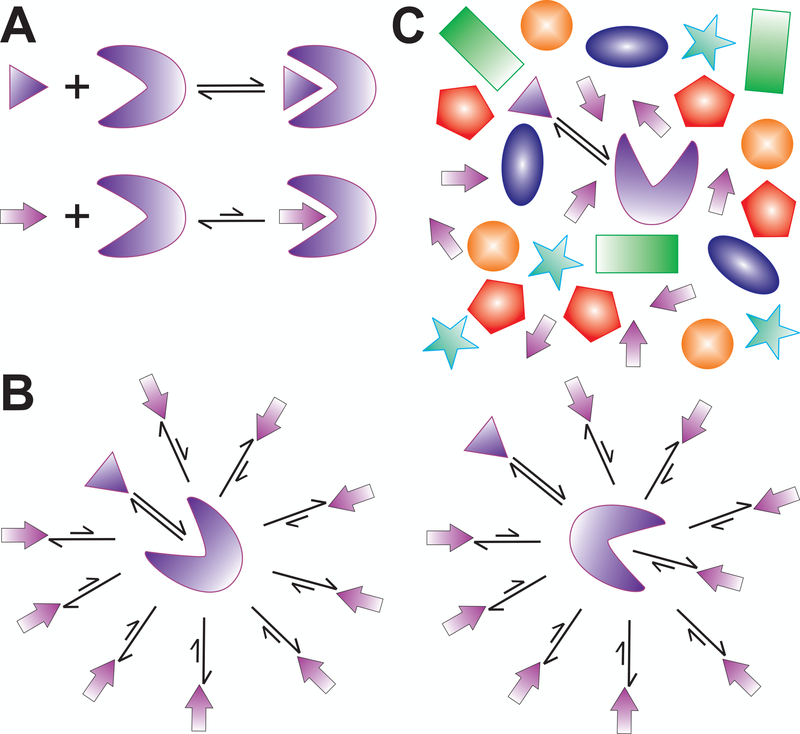
Now suppose that S is significantly more specific for binding L than competitor C, manifested in its Kd,S being 1,000-fold tighter (i.e., smaller) than that of C, termed Kd,C. Yet, if C were either found at 1,000-fold higher cellular concentration than S, or there existed 1,000 different forms of competitor C (C1, C2, C3, …), each at the same low concentration of S, then we would end up with the same scenario of L being occupied by the specific binding partner S only 33% of the time, rather than 50%. That is, based on competition with a plethora of weak binders, the law of mass action – in the form of a set of coupled molecular equilibria – predicts that the concentration of a target available to a tight binder is lowered, shifting the equilibrium per Le Chatelier away from high-specificity binding (Fig. 2A,B). Importantly, these fundamental considerations are similarly at work in a more complex and realistic scenario where the concentrations and affinities of the participating components are more diverse than assumed in the above back-of-the-envelope example.
Figure 2.
The consequences of competition among a plethora of cellular components. Reversible equilibrium binding of a high-affinity (top) and low-affinity (bottom) partner to a biopolymer of interest (A) becomes shifted by Le Chatelier towards the low-affinity binder when that is at comparably high concentration (B). In the crowded molecular environment of cell, this shift if even more significant due to the high number of diverse, weak competitors (C).
3. The Cell’s Reality is Diversely Crowded
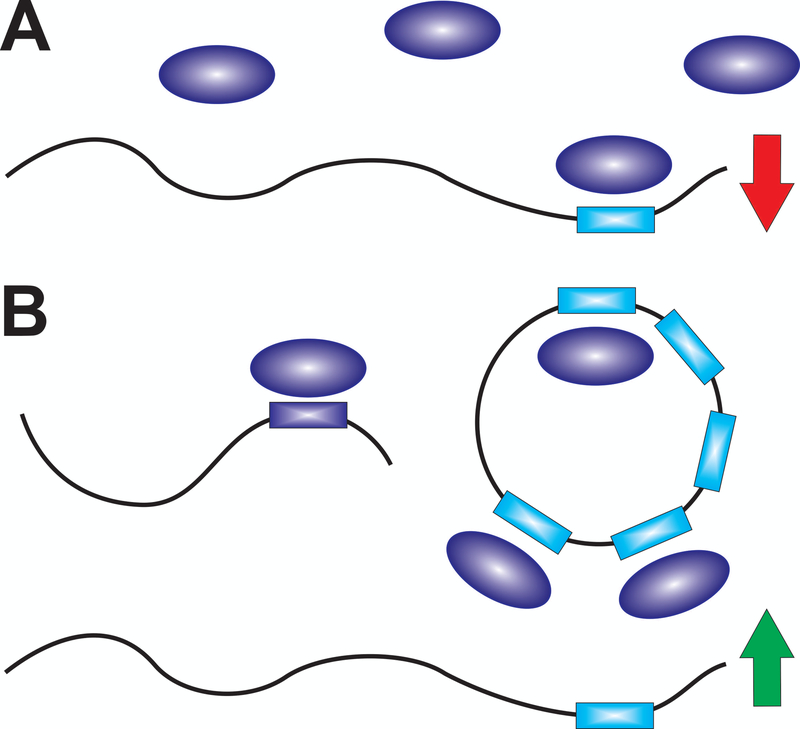
The predicted significant impact of a large number of low-affinity competitors on specific binding interactions is the reality of the cell’s crowded molecular environment (Fig. 2C). In fact, increasing evidence is emerging that, for example, many cryptic RNA-binding sites exist on proteins of all trades, including many metabolic housekeeping enzymes.[3,22,23] Similarly, such dynamic equilibrium considerations are the essence of recent modeling efforts of the regulatory RNA silencing machinery, whereby each RNA target competes both for protein partners of the RNA induced silencing complex (RISC) and microRNAs (miRNAs; Fig. 3).[24–26] This competition can lead to some non-intuitive behaviors, such as that highly expressed miRNAs become sequestered and thus neutralized in terms of their regulatory power by targets that ‘sponge’ them up, whereas competition between binding sites on different RNAs can lead to regulatory crosstalk and even apparent upregulation of targets (Fig. 3B).[24,25] Importantly, the severity of these effects depends on both the affinities of all potential binding partners and the extent to which a specific component(s) of a given pathway becomes limiting in concentration, which the cell of course can control dynamically through gene regulation. Notably, evolution will fine-tune the affinity and gene expression level of each cellular component concomitantly, leading to highly complex networks of interactions where all content of the cell is interconnected and some components become particularly highly connected system hubs. Spontaneous evolutionary events such as a gene duplication or mutation in a binding interface will therefore not only affect the immediate pathway in which a cellular component is majorly involved, but also any other strong or weak interaction the component has the capacity to engage within the cell. This is particularly true since the cell is only (tens of) micrometers in size; barrier-less molecular diffusion from one end to the other is thus generally fast (sub-second time scales) so that all cellular components can in principle encounter each other continuously to realize all of these network interactions.
Figure 3.
Known examples for equilibrium binding competition affecting biological function. During RNA silencing (A), an activated RISC (dark blue oval) recognizes a miRNA response element (MRE, cyan rectangle) on an mRNA to downregulate its expression (red arrow). Upon expression of a significant concentration of an mRNA with an MRE of higher affinity (dark blue rectangle), or of a circular RNA with multiple MREs, a targeted mRNA will seemingly be upregulated (green arrow) due to the shifting coupled equilibrium siphoning off the limiting RISC (B).
But this picture raises the puzzling question - If crowding in a small volume and a plethora of interactions of varying strength render everything in a cell part of one large, interconnected network, what then specifies cellular pathways? For one, our linear cause-and-effect expectation bias arguably makes us “find” pathways everywhere, while in reality networking aspects are more pervasive than typically considered. Second, cells have developed ways to suppress undesired interactions, for example, by compartmentalization through barriers such as membranes or separated liquid-liquid phases,[27–29] or by dynamic (often reversible) post-biosynthetic modifications of interaction interfaces, i.e., post-translational modifications of a protein,[30] post-transcriptional modifications of an RNA (the “epitranscriptome”),[27,31,32] or epigenetic modifications of a DNA[33]. Such spatiotemporal organization helps shape the network into “funnels” (or, mathematically, “attractors”) that the cell can remodel on demand, essentially by modulating the active local concentration of (potential) network interaction partners. Third, and perhaps most importantly, biology (or evolution) fine-tunes the baseline affinities of active pathway components in order to strengthen subsets of connectivities within the network so that specific pathways arise from the “forest”; conversely, non-productive interactions will be selected against.[34]
Yet, even well-studied metabolic pathways have to be amended and expanded regularly, with important implications for normal physiology and diseases such as cancer.[35–38] Similarly, recent studies have found that hundreds of proteins and RNAs “moonlight” in that they each participate in multiple distinct biochemical pathways by using different parts of their surfaces to interact with diverse binding partners.[3,22,23,39,40] When even metabolic enzymes that are thought to “normally” phosphorylate small metabolites equally function as protein kinases and phosphorylate a variety of protein substrates to regulate fundamental cellular functions,[41] it is no wonder that cellular networks show much more redundancy and plasticity, and are harder to therapeutically target, than expected from simple linear cause-and-effect relationships.
4. Kinetic Proofreading, Partitioning, and Trapping
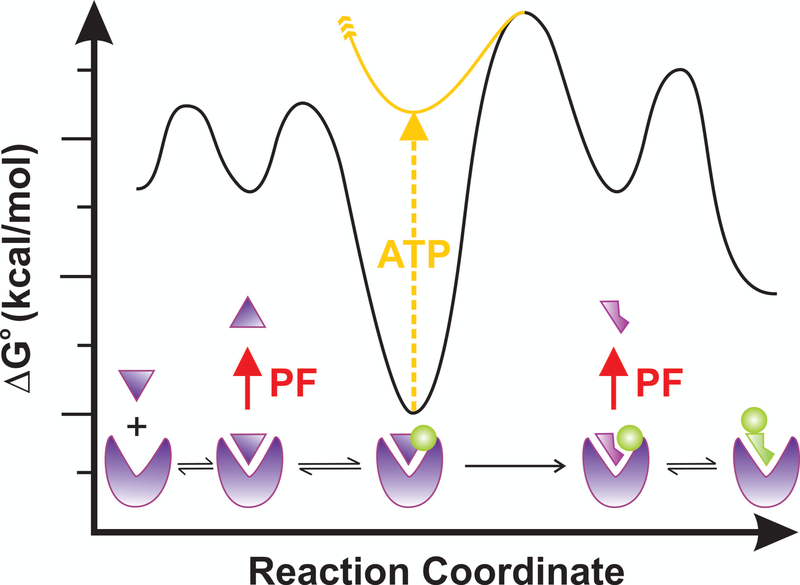
If thermodynamics and the law of mass action represent the “Yin” underlying biology, kinetics is the inseparable (and sometimes contradictory) “Yang.” In fact, thermodynamics may not be the most important force in biology because, per Heraclitus of Ephesus, “Life is Flux” (or in Greek, “Panta Rhei,” meaning that change is the only constant in life).[42] Accordingly, systems biology uses dynamic flux balance analysis as a mathematical approach to understand the flow of metabolites through a metabolic reaction network.[43] Most individual reactions within metabolic pathways are reversible except for a few (near-)irreversible steps that oftentimes occur early in the pathway and are highly regulated (Fig. 1). Similarly, complex biological processes as diverse as mRNA translation,[44–47] pre-mRNA splicing,[48,49] nucleotide excision repair of damaged DNA,[50] and discrimination of self- and non-self-peptides by T cells[51] all rely on a mix of sequential reversible and irreversible steps to effect what is called “kinetic proofreading” (Fig. 4). As the name implies, this mechanism enhances biological specificity and fidelity beyond the level that can ordinarily be achieved by the difference in thermodynamic free energy between correct and incorrect interactions.[52,53] In particular, the insertion of energy-consuming and therefore (near-)irreversible (or unidirectional) steps isolates the flanking reversible steps from one another so that kinetic partitioning between moving forward and instead going off-path and disassembling the complex can occur independently, both before and after the irreversible step (Fig. 4), thus in effect multiplying the two respective error frequencies for enhanced fidelity. In fact, even without irreversible consumption of energy some level of “conformational proofreading” is achieved by a programmed structural mismatch between two binding partners that have to strain (i.e., undergo an induced fit) to match each other perfectly.[54] For example, the ribosome is thought to have evolved into an optimal decoder by using the binding energy of an optimally decoding tRNA for conformational distortions that distinguish it from a multitude of near-cognate substrates.[55]
Figure 4.
The basis of kinetic proofreading. An enzyme reversibly binds its substrate that can be rejected (red arrow) or further accommodated upon binding of a cofactor (green circle, “pawl”). Upon consumption of energy through ATP hydrolysis or some other energetic process, the free energy (ΔG) landscape changes in ways that allow for a (near-)irreversible step towards a new conformation or configuration, wherein another, independent rejection step completes the proofreading (PF) against non- or near-cognate substrates. Only if the kinetics of the forward steps exceed that of the rejection steps will the reaction be completed successfully.
Taken together, the cell is a cauldron churning far from equilibrium where the “magic” of life emerges with the help of energy-consuming reactions that make certain steps kinetically irreversible and so help specify the pathways currently favored among the entire network. The network – like any “safety net” – provides added stability and robustness in the face of outside stress; yet it also enables dynamic system control through feedback and feedforward loops[56] that lead to “emergent” cellular behaviors that we identify as “life,” including homeostasis, responsiveness, adaptation, growth, reproduction and heredity. Notably, if thermodynamics were dominating, a low-energy (stable) state would always end up kinetically trapping each pathway, without (much) further change. Instead, Nature utilizes kinetic traps as “attractors” along a pathway but then makes them shorter-lived by utilizing external energy (often in the form of NTP hydrolysis, but also other chemical conversions) to (re)move a trapping component (or “pawl”; Fig. 4). The resulting “biased Brownian ratchet machine” draws path directionality from the random thermal fluctuations it constantly experiences but then directionally “rectifies” or “biases” with the energy-consuming irreversible step.[49,57,58]
Specificity within biological pathways therefore has both thermodynamic and kinetic drivers. In light of the onslaught of competing non-specific, weak interactions, on-pathway specific interactions will be those both thermodynamically sufficiently stable and kinetically sufficiently long-lived to survive and not be discarded or proofread. Furthermore, since the kinetic behavior of each single molecule in the cell is highly stochastic, with the individual event onset (or wait) times often exponentially distributed,[59] the order in which steps occur in a complex pathway can sometimes vary, as was observed, for example, during spliceosome assembly.[60] Such stochasticity results in the co-existence of multiple parallel pathways, all productive, and is one manifestation of so-called “fuzzy logic” in biology, where the binary all-or-none choice of computer logic is replaced with partial truth values ranging from completely true to completely false.[61,62]
5. Biological Specificity, Refined by Diverse Cellular Interactions
Perhaps a quintessential example of the challenges associated with transferring the concept of specificity from purified in vitro systems to the cellular environment is the debate of Mg2+ homeostasis.[63,64] Only recently – late compared to the better understood, chemically similar earth alkali metal ion Ca2+ (Refs. [65–67]) – has it become clear that the cell actively maintains a high (millimolar) Mg2+ concentration using specialized ion channels.[64,68–70] The exact concentration of free Mg2+ in the cell is hotly debated and varies with the KD of the detection probe, with this KD itself being cell-condition (pH, ionic strength etc.) dependent, which has led to varying estimates of the free Mg2+ concentration ranging between 0.5–2 mM.[71] What sometimes seems forgotten, however, is that a large fraction of the total ~20 mM Mg2+ in the cell is bound by nucleic acids, especially RNA, since ~90% of the negatively charged phosphates are charge-neutralized by Mg2+;[72] similarly, each NTP typically chelates one Mg2+ ion.[73] Given its rapid dissociation rate constant, the divalent ion therefore will quickly equilibrate between many Mg2+-binding sites in the cell (Fig. 3) so that a chelating detection probe is subject to competition for binding of Mg2+, especially from those competitors with a higher affinity for Mg2+ or a higher concentration than the probe itself (Fig. 2). An example is the recent observation that amino acid chelated Mg2+ pools are readily functional in promoting RNA folding and function[74]. What fraction of Mg2+ is detected as “free” will therefore be extremely sensitive to both the probe used and all diverse molecular species present in the cell at the time, as well as the molecular characteristics of their individual Mg2+ binding sites. It is no wonder that much ambiguity arises from such measurements.
Another instructive example is that of transcription initiation in the eukaryotic cell, a very dynamic process driven by promoters, transcription factors, and distal enhancers.[75] These elements jointly recruit RNA polymerase to initiate transcription on specific start sites in a highly reversible assembly process wherein transcription factors rapidly exchange between possible binding sites; in effect, promoters are constantly competing for both cofactors and RNA polymerase.[75] To facilitate this process, many eukaryotic transcription factors contain intrinsically disordered, so-called low-complexity domains (LCD) that allow them to rapidly form local high-concentration hubs through dynamic, multivalent, specific LCD-LCD interactions that recently have been visualized by live-cell single molecule imaging.[76,77] These observations are an example of the rapidly expanding universe of functionally important, dynamic liquid-liquid phase separation (LLPS) processes that make cellular life more efficient by organizing it into membrane-less organelles of enhanced local concentration.[27–29] Transcription initiation is thus one exemplary process where the cell both exploits and suppresses the law of mass action inside and outside, respectively, of a condensed liquid phase in dynamic fashion. Even more fundamentally, the competition between strongly binding promoter sequences and all other segments of a DNA weakly associating with an RNA polymerase undoubtedly ensures that only highly cooperative, multivalent transcription initiation complexes will result in successful commitment to gene expression[78]. Similarly, it has been shown that the empirically observed “Savageau demand rules” that genes needed often tend to be regulated by transcription activators, whereas genes rarely needed are more often regulated by repressors, can be explained by the fact that in both cases the activator or repressor site is occupied most of the time, thus minimizing errors induced by weakly binding competitors.[79,80]
Our considerations so far assumed effectively time-invariant pairwise interactions between binding or reaction partners, modulated by direct competition with other pairwise interactions. An additional dimension often absent from or not considered in purified biophysical systems are secondary effects arising from higher-order interactions. A classic example are allosteric effects where the binding of one interaction partner modulates the binding affinity towards another partner, and vice versa, such as allosteric activation or inhibition of a protein enzyme.[81,82] For biological machines such as the ribosome[83] or the spliceosome[84] such allosteric effects are known to facilitate an ordered assembly and reaction process. And yet, they are significantly beyond simple cause-and-effect relationships, with thousands of different molecules in a cell undoubtedly having a plethora of allosteric effects on each other that still await discovery, further increasing interconnectivity and plasticity. Given their transient nature and unknown functional consequences, we probably will still need to develop entirely new screening tools to be able to map these dynamic interconnections in their entirety.
6. Outlook and Call for Action
As we continue to peer deeper and deeper into the cauldron of cellular life we are starting to understand more of its sophisticated spatiotemporal organization where the real estate agents’ mantra of “location, location, location” is critical,[85] yet of course is only a consequence of specific “interactions, interactions, interactions”. Understanding the specificity of these interactions is at the heart of deciphering the deeper “code of life”,[86] a challenge that runs counter to human intuition but deserves our full attention. To meet this challenge, we need to acknowledge the fact that, just as in societal life, diversity (in affinity and concentration) matters since it profoundly shapes biomolecular specificity in often unexpected ways by enhancing competition. And just as in modern societies, diversity in biology creates both challenges and opportunities. Recent progress in moving biophysical inquiries into the cell[15,87,88] gives hope that we are at the beginning of truly unearthing the foundations of biology. More tools for direct intracellular probing of biological pathways still need to be developed, including – but not limited to – improved (higher-contrast, higher-resolution, more versatile) fluorescent and other minimally invasive imaging probes, more in-cell manipulation tools (such as genome editing and synthetic biology circuitry), elegant ways to combine biochemical fractionation of cellular components with tagging (barcoding) their spatiotemporal origins in the cell, multiplexed and high-throughput molecular characterization tools for cellular components probed in situ, and more powerful systems biology tools to simulate ever more complex interaction networks. As always, new tools will undoubtedly become drivers for discovering new biology, ultimately solving the question of how biological pathways breathe life into a cell.
Acknowledgments
N.G.W. appreciates the thorough reading and valuable feedback provided by Alexander Johnson-Buck, acknowledges support from NIH R01 grants GM062357, GM118524 and GM122803, and is thankful for helpful suggestions from several anonymous reviewers.
Footnotes
Conflict of Interest
The author declares no commercial or financial conflict of interest.
References
- [1].Pauling L, Marinacci B. 1995. Linus Pauling in His Own Words: Selections From His Writings, Speeches and Interviews New York City: Simon and Schuster. [Google Scholar]
- [2].Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Nature 2012, 489, 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Cell 2012, 149, 1393–406. [DOI] [PubMed] [Google Scholar]
- [4].Belinky F, Bahir I, Stelzer G, Zimmerman S, Rosen N, Nativ N, Dalah I, Iny Stein T, Rappaport N, Mituyama T, Safran M, Lancet D. Bioinformatics 2013, 29, 255–61. [DOI] [PubMed] [Google Scholar]
- [5].Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segre AV, Djebali S, Niarchou A, Consortium GT, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigo R. Science 2015, 348, 660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJ, Arner E, Hayashizaki Y, Carninci P, Forrest AR. Nature 2017, 543, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Science 2015, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- [8].Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, Backstrom A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjostedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Ponten F, von Feilitzen K, Lilley KS, Uhlen M, Lundberg E. Science 2017, 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- [9].Palazzo AF, Lee ES. Front. Genet 2015, 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mudge JM, Harrow J. Nat. Rev. Genet 2016, 17, 758–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, Thorleifsson SG, Agren R, Bolling C, Bordel S, Chavali AK, Dobson P, Dunn WB, Endler L, Hala D, Hucka M, Hull D, Jameson D, Jamshidi N, Jonsson JJ, Juty N, Keating S, Nookaew I, Le Novere N, Malys N, Mazein A, Papin JA, Price ND, Selkov E Sr., Sigurdsson MI, Simeonidis E, Sonnenschein N, Smallbone K, Sorokin A, van Beek JH, Weichart D, Goryanin I, Nielsen J, Westerhoff HV, Kell DB, Mendes P, Palsson BO. Nat. Biotechnol 2013, 31, 419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Noble D. 2006. The Music of Life Oxford: Oxford University Press. [Google Scholar]
- [13].Xie XS, Yu J, Yang WY. Science 2006, 312, 228–30. [DOI] [PubMed] [Google Scholar]
- [14].Li GW, Xie XS. Nature 2011, 475, 308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pitchiaya S, Heinicke LA, Custer TC, Walter NG. Chem. Rev 2014, 114, 3224–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Stockdale FE, Quake SR, Pease RF, Mindrinos MN, Bhanot G, Dairkee SH, Davis RW, Jeffrey SS. PLoS One 2012, 7, e33788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Swanton C, Govindan R. N. Engl. J. Med 2016, 374, 1864–73. [DOI] [PubMed] [Google Scholar]
- [18].Waage P, Guldberg CM. 1864. Studies of affinities: Forhandlinger. [Google Scholar]
- [19].van’t Hoff JH. Ber. Deut. Chem. Gesell. Berlin 1877, 10, 669–78. [Google Scholar]
- [20].Dasmeh P, Searles DJ, Ajloo D, Evans DJ, Williams SR. J Chem Phys 2009, 131, 214503. [DOI] [PubMed] [Google Scholar]
- [21].Samuelson PA. 1983. Foundations of Economic Analysis: Harvard University Press. [Google Scholar]
- [22].Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. Mol. Cell 2016, 63, 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Daher M, Widom JR, Tay W, Walter NG. J. Mol. Biol 2018, 430, 509–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nyayanit D, Gadgil CJ. RNA 2015, 21, 307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jens M, Rajewsky N. Nat. Rev. Genet 2015, 16, 113–26. [DOI] [PubMed] [Google Scholar]
- [26].Gurtan AM, Sharp PA. J. Mol. Biol 2013, 425, 3582–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Drino A, Schaefer MR. Bioessays 2018, e1800085. [DOI] [PubMed] [Google Scholar]
- [28].Wheeler RJ, Hyman AA. Philos. Trans. R Soc. Lond. B Biol. Sci 2018, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Langdon EM, Gladfelter AS. Annu. Rev. Microbiol 2018, 72, 255–71. [DOI] [PubMed] [Google Scholar]
- [30].Prabakaran S, Lippens G, Steen H, Gunawardena J. Wiley Interdiscip. Rev. Syst. Biol. Med 2012, 4, 565–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Helm M, Y. Motorin. Nat. Rev. Genet 2017, 18, 275–91. [DOI] [PubMed] [Google Scholar]
- [32].Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. RNA 2017, 23, 1754–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goldberg AD, Allis CD, Bernstein E. Cell 2007, 128, 635–8. [DOI] [PubMed] [Google Scholar]
- [34].Yang JR, Liao BY, Zhuang SM, Zhang J. Proc. Natl. Acad. Sci. USA 2012, 109, E831–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, Heggermont W, Godde L, Vinckier S, Van Veldhoven PP, Eelen G, Schoonjans L, Gerhardt H, Dewerchin M, Baes M, De Bock K, Ghesquiere B, Lunt SY, Fendt SM, Carmeliet P. Nature 2015, 520, 192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Levin E, Lopez-Martinez G, Fane B, Davidowitz G. Science 2017, 355, 733–5. [DOI] [PubMed] [Google Scholar]
- [37].Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, White E, Rabinowitz JD. Nature 2017, 551, 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, Li H, Huet G, Yuan Q, Wigal T, Butt Y, Ni M, Torrealba J, Oliver D, Lenkinski RE, Malloy CR, Wachsmann JW, Young JD, Kernstine K, DeBerardinis RJ. Cell 2017, 171, 358–71 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Docter BE, Horowitz S, Gray MJ, Jakob U, Bardwell JC. Nucleic Acids Res. 2016, 44, 4835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jeffery CJ. Philos. Trans. R Soc. Lond. B Biol. Sci 2018, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lu Z, Hunter T. Trends Biochem. Sci 2018, 43, 301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mark JJ. 2010. Heraclitus_of_Ephesus. Anc. Hist. Encyclop [Google Scholar]
- [43].Orth JD, Thiele I, Palsson BO. Nat. Biotechnol 2010, 28, 245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Spirin AS. Biochemistry 2009, 48, 10688–92.19835415 [Google Scholar]
- [45].Frank J Jr., Gonzalez RL. Annu. Rev. Biochem 2010, 79, 381–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rodnina MV, Wintermeyer W. Biochem. Soc. Trans 2011, 39, 658–62. [DOI] [PubMed] [Google Scholar]
- [47].Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nat. Struct. Mol. Biol 2004, 11, 1008–14. [DOI] [PubMed] [Google Scholar]
- [48].Koodathingal P, Staley JP. RNA Biol. 2013, 10, 1073–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. Nat. Struct. Mol. Biol 2013, 20, 1450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hu J, Selby CP, Adar S, Adebali O, Sancar A. J. Biol. Chem 2017, 292, 15588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jansson A. Self Nonself 2011, 2, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hopfield JJ. Proc. Natl. Acad. Sci. USA 1974, 71, 4135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ninio J. Biochimie 1975, 57, 587–95. [DOI] [PubMed] [Google Scholar]
- [54].Savir Y, Tlusty T. PLoS One 2007, 2, e468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Savir Y, Tlusty T. Cell 2013, 153, 471–9. [DOI] [PubMed] [Google Scholar]
- [56].Kitano H. Science 2002, 295, 1662–4. [DOI] [PubMed] [Google Scholar]
- [57].Cordova NJ, Ermentrout B, Oster GF. Proc. Natl. Acad. Sci. USA 1992, 89, 339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Astumian RD. Science 1997, 276, 917–22. [DOI] [PubMed] [Google Scholar]
- [59].Moffitt JR, Chemla YR, Bustamante C. Methods Enzymol. 2010, 475, 221–57. [DOI] [PubMed] [Google Scholar]
- [60].Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Correa IR Jr., Xu MQ, Gelles J, Moore MJ. Cell Rep. 2013, 5, 151–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zadeh LA. Inform. Control 1965, 8, 338–53. [Google Scholar]
- [62].Novák V, Perfilieva I, Močkoř J. 1999. Mathematical principles of fuzzy logic Dodrecht: Kluwer Academic. [Google Scholar]
- [63].Johnson-Buck AE, McDowell SE, Walter NG. Met. Ions Life Sci. 2011, 9, 175–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Romani AM. Arch. Biochem. Biophys 2011, 512, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Berridge MJ, Lipp P, Bootman MD. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [DOI] [PubMed] [Google Scholar]
- [66].Berridge MJ, Bootman MD, Roderick HL. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–29. [DOI] [PubMed] [Google Scholar]
- [67].Clapham DE. Cell 2007, 131, 1047–58. [DOI] [PubMed] [Google Scholar]
- [68].Wolf FI. Sci. STKE 2004, 2004, pe23. [DOI] [PubMed] [Google Scholar]
- [69].Wolf FI, V. Trapani. Magnes. Res 2011, 24, S86–91. [DOI] [PubMed] [Google Scholar]
- [70].de Baaij JH, Hoenderop JG, Bindels RJ. Physiol. Rev 2015, 95, 1–46. [DOI] [PubMed] [Google Scholar]
- [71].T. Gunther. Magnes. Res 2006, 19, 225–36. [PubMed] [Google Scholar]
- [72].Porschke D, Burke JM, Walter NG. J. Mol. Biol 1999, 289, 799–813. [DOI] [PubMed] [Google Scholar]
- [73].Pecoraro VL, Hermes JD, Cleland WW. Biochemistry 1984, 23, 5262–71. [DOI] [PubMed] [Google Scholar]
- [74].Yamagami R, Bingaman JL, Frankel EA, Bevilacqua PC. Nat Commun 2018, 9, 2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Haberle V, A. Stark. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, Tjian R. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, Abraham BJ, Afeyan LK, Guo YE, Rimel JK, Fant CB, Schuijers J, Lee TI, Taatjes DJ, Young RA. Cell 2018, 175, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tafvizi A, Mirny LA, van Oijen AM. Chemphyschem 2011, 12, 1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Shinar G, Dekel E, Tlusty T, Alon U. Proc Natl Acad Sci U S A 2006, 103, 3999–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sasson V, Shachrai I, Bren A, Dekel E, Alon U. Mol Cell 2012, 46, 399–407. [DOI] [PubMed] [Google Scholar]
- [81].Hilser VJ, Wrabl JO, Motlagh HN. Annu. Rev. Biophys 2012, 41, 585–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Motlagh HN, Wrabl JO, Li J, Hilser VJ. Nature 2014, 508, 331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Steitz TA. Nat. Rev. Mol. Cell Biol. 2008, 9, 242–53. [DOI] [PubMed] [Google Scholar]
- [84].Fica SM, Nagai K. Nat. Struct. Mol. Biol 2017, 24, 791–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hurtley S. Science 2009, 326, 1205. [DOI] [PubMed] [Google Scholar]
- [86].Collins FS, Jegalian KG. Sci. Am 1999, 281, 86–91. [DOI] [PubMed] [Google Scholar]
- [87].Plitzko JM, Schuler B, Selenko P. Curr. Opin. Struct. Biol 2017, 46, 110–21. [DOI] [PubMed] [Google Scholar]
- [88].Giassa IC, Rynes J, Fessl T, Foldynova-Trantirkova S, Trantirek L. FEBS Lett. 2018, 592, 1997–2011. [DOI] [PubMed] [Google Scholar]