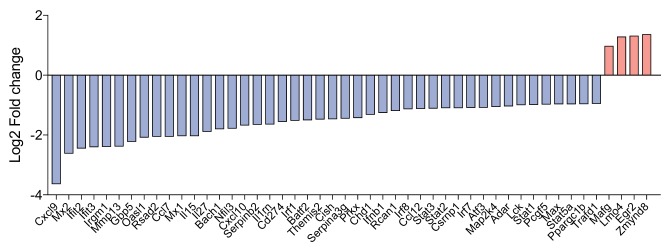
Figure 2. Regulator genes are induced above a threshold of TLR4 ligand.
(A, B) Using the titration matrix conditions shown in Figure 1, BMDM were stimulated for 2 hr and induced gene expression was measured by microarray. Differential gene expression analysis was performed between the hotspot and TLR4 high regions of the dual ligand titration matrix. Candidate regulator genes preferentially induced by high TLR4 ligand are noted (B). (C) BMDM were treated with three independent siRNA per candidate regulator (siRNA resulting in less than 50% viability were excluded), MyD88 siRNA, or non-targeting control (NTC) siRNA for 48 hr prior to stimulation with 1000 nM P3C and 100 nM KLA (TLR4hi condition). After 6 hr of stimulation, TNF or CXCL1 was quantified in supernatants by ELISA and values were normalized based on cell viability. Individual data points represent wells treated independently with siRNA and TLR ligand, and the gray bar depicts NTC standard deviation. Candidate genes were differentially expressed in three independent microarray or qPCR experiments and siRNA results are representative of two independent experiments. Stars represent statistical significance based on ordinary one-way ANOVA, versus control (*p≤0.05). (D) BMDM were stimulated with the indicated concentrations of TLR ligand and harvested at the indicated time. Gene expression was quantified using qPCR and fold change was calculated compared to unstimulated BMDM. Data are representative of three independent experiments.