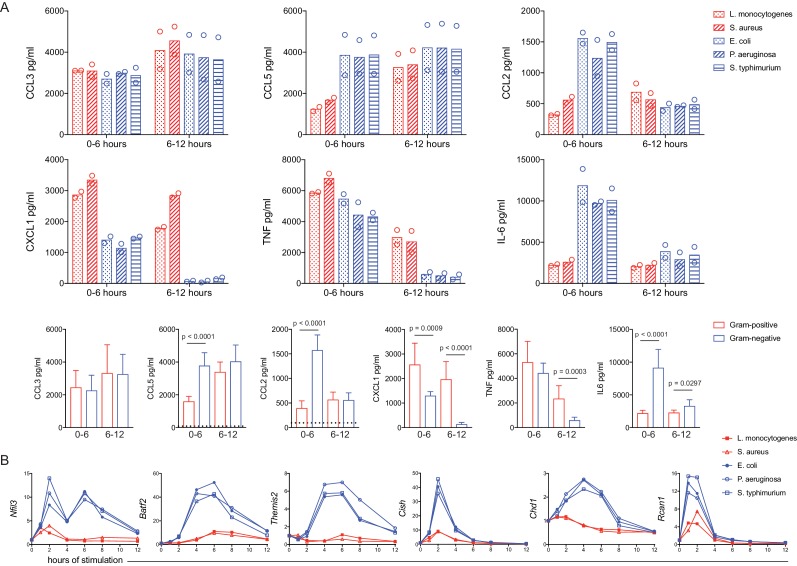
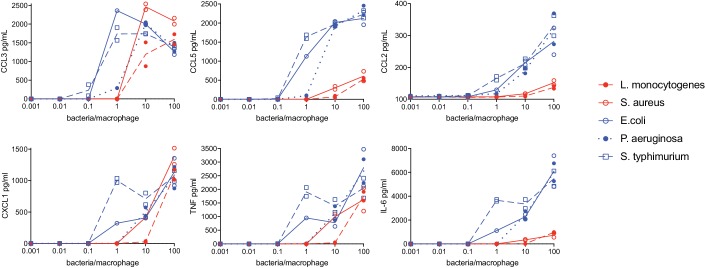
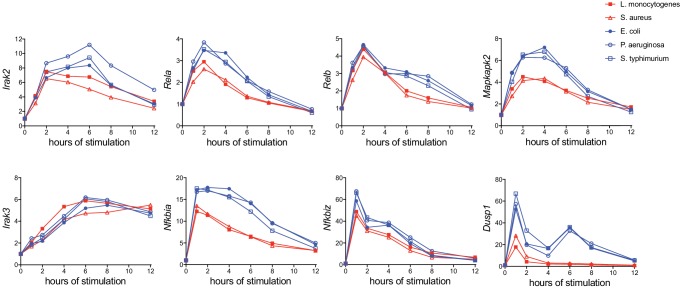
Figure 3. A dual TLR titration matrix predicts bacteria-specific cytokine shutdown and regulator induction.
BMDM were stimulated with heat-killed bacteria. (A) After 6 hr of stimulation, supernatants were removed (0–6 hr) and replaced with fresh medium for a subsequent 6 hr of stimulation (6–12 hr). Cytokines were quantified using cytometric bead array and data points are experimental replicates. Data from bacterial species (upper rows) were combined from three independent experiments (lower row) and statistical significance was determined using unpaired t-tests between Gram-negative- and Gram-positive-stimulated samples; where no p-value is indicated, results were not significant. Dotted lines in the lower row represent the mean values for unstimulated BMDM (0–12 hr). In some cases, these controls are not visible because they are below the level of detection (a zero value). (B) Cells were harvested at the indicated time, gene expression was quantified using qPCR, and fold change was calculated compared to unstimulated BMDM. Results are representative of four or more independent experiments.