Abstract
Over the past decade, photoswitchable molecules have been emerging as attractive tools for investigating biological processes with spatiotemporal resolution in a minimally invasive fashion. Photoswitches built on light-sensitive proteins or domains have significantly advanced neuronal and cellular studies. To install photosensitivity to general proteins and to enable high specificity for modulation, photoswitchable click amino acids (PSCaas) based on azobenzene have been developed and recently genetically incorporated into proteins via the expansion of the genetic code. PSCaas allow targeting selected sites in a protein for high specificity and are generally applicable to various proteins. In addition, PSCaas contain a click functional group, which selectively reacts with an appropriately positioned cysteine forming a photocontrollable bridge on the protein in situ. The photocontrollable bridge enables reversible modulation of the secondary structure of the spanned region and thus the function of the protein. In this chapter we describe the design and genetic encoding of PSCaa. Protocols are presented for incorporating PSCaa into a model protein calmodulin to build the bridge followed by photocontrol of calmodulin’s conformation and binding function.
1. INTRODUCTION
Various optical techniques have been developed to alter biological function and gain information about physiological processes. A prominent method is optogenetics, taking advantage of the photoactivation of opsin proteins embedded in the membrane of neurons to manipulate neuronal activity by light-directed interference (Fenno, Yizhar, & Deisseroth, 2011). Optogenetics revolutionized the neurosciences allowing researchers to shed light on the role of ion channels in neuronal activity (Bernstein & Boyden, 2011). Despite its important role in neuroscience, its pathway to become a general tool for the photocontrol of protein function and activity is limited. In recent years, other light-sensitive domains (e.g. LOV = Light Oxygen Voltage) have been used to trigger biological events (Pudasaini, El-Arab, & Zoltowski, 2015). Fusing LOV domains to cytosolic proteins allows for an optical control which is not limited to the membrane environment. However, those domains are large which may cause interferences (e.g. aggregation), and they hardly allow investigation of a specific site or controlling conformational states. The use of small molecule photoswitches can mitigate these drawbacks. Using small molecule photoswitches, biological activity can be photocontrolled by either caging critical residues of the protein with photoremovable protecting groups or by attaching photoisomerizable units (Brieke, Rohrbach, Gottschalk, Mayer, & Heckel, 2012; Szymański, Beierle, Kistemaker, Velema, & Feringa, 2013).
Although caged amino acids can be introduced into proteins in vivo using modern unnatural amino acid (Uaa) mutagenesis techniques, these Uaas cannot afford reversible control (Kang et al., 2013; Deiters, Groff, Ryu, Xie, & Schultz, 2006). In addition, photodecaging can control a single residue only to induce localized perturbations. Biological function often requires the change of protein domains in structure and conformation, which cannot be readily achieved by targeting a single residue (Courtney & Deiters, 2018). Photoswitches bridging two residues can provide a general approach to photocontrol protein conformation (Beharry & Woolley, 2011; Zhang et al., 2009). Such a photoswitch can be introduced into proteins by connecting two appropriately located cysteines with an azobenzene unit bearing two thiol reactive anchoring groups. However, the anchoring groups can react with any thiol in the surrounding milieu, resulting in undesired products and thus preventing the use of this method in live cells.
To overcome this limitation and to install photocontrollable bridges in general proteins, PhotoSwitchable Click Amino Acids (PSCaa) were developed and site specifically incorporated into proteins via the expansion of the genetic code (Fig 1A) (Hoppmann et al., 2014; Hoppmann, Schmieder, Heinrich, & Beyermann, 2011). An orthogonal tRNA/synthetase pair evolved to be specific for the PSCaa allows for site-specific incorporation of the PSCaa into proteins in response to a unique codon (e.g., the amber stop codon, UAG) using the translational machinery. In contrast to phenylazophenylalanine 1 that was previously genetically incorporated (Bose, Groff, Xie, Brustad, & Schultz, 2006), PSCaas 2–4 consist of a photoisomerizable azobenzene side chain as well as a thiol reactive group, which enables covalent bonding with an appropriately positioned cysteine within a protein domain, forming the photocontrollable bridge in situ. The resultant bridge allows light-inducible control of protein conformation and, by extension, its activity (Hoppmann et al., 2014; Hoppmann, Maslennikov, Choe, & Wang, 2015) (Fig 1B). In this chapter, we discuss the genetic encoding of PSCaa to control the conformation and activity of proteins.
Figure 1.
(A) Structures of the photoswitchable click amino acids and phenyalzoazophenylalanine amino acid 1. PSCaa 2: 2-amino-3-(4-((3-vinylphenyl)diazenyl)phenyl)propanoic acid; Cl-PSCaa 3: 2-amino-3-(4-((3-(chloromethyl)phenyl)diazenyl)phenyl)propanoic acid; F-PSCaa 4: (S,E)-2-amino-3-(4-((pentafluorophenyl)-diazenyl)phenyl)propanoic acid; Keto-PSCaa 5: 2-amino-3-(4-((3-(2-acetyl)phenyl)diazenyl)phenyl)propanoic acid. (B) Concept of photoswitchable click amino acid shown for F-PSCaa: The genetic incorporation of PSCaa into a protein domain bearing a cysteine at an appropriate site allows the formation of a photocontrollable bridge on the target protein in situ.
2. Genetic incorporation and application of photoswitchable click amino acids
We will first discuss the design of the PSCaa, followed by a brief description of evolving aminoacyl-tRNA synthetases specific for PSCaas and the incorporation of PSCaas into model proteins. We will then summarize a few ways for detecting the photoisomerization of PSCaa and its bridge. Finally, we will briefly discuss biological applications of the genetically encoded PSCaas.
2.1. Design of photoswitchable click amino acid (PSCaa)
The idea of using PSCaa to control conformation and activity was initially demonstrated using a model peptide, where PSCaa 2 had an alkene unit to allow for light-induced thiol-ene click reaction with appropriately positioned cysteine residues (Hoppmann, Kühne, & Beyermann, 2012; Hoppmann et al., 2011). By chemically synthesizing PSCaa 2 and placing a cysteine at appropriate site (e.g. i+4 helical distance) into the peptide hormone urocortin, a photocontrollable bridge similar to a staple was generated that allowed the conformation of the urocortin to be directed by light, and thus the binding of the hormone to the CRF receptor.
Over the years, the design of PSCaa was advanced such that the covalent bond formation was enabled in situ without using toxic UV light to induce the thiol-ene click chemistry. The Cl-PSCaa 3 contains a benzyl chloride as a thiol-reactive handle, which reacts with a nearby cysteine spontaneously via a proximity-enabled nucleophilic substitution. The benzyl chloride in Cl-PSCaa is reactive to cysteine, but stable enough to be used in biological matrices where excess of thiols is present (e.g. glutathione). Therefore, Cl-PSCaa 3 has been genetically incorporated into proteins using orthogonal tRNA/synthetase in E. coli and mammalian cells (Hoppmann et al., 2014).
Another advancement is the design of PSCaas in response to light of longer wavelength. PSCaa and Cl-PSCaa are azobenzene type photoswitches showing E-to-Z photoisomerization upon UV light activation, generating a photostationary equilibrium (or photostationary state, pss) in which the Z-isomer is enriched. To design a visible light activatable PSCaa, we recently installed a fluorinated phenyl ring that not only allowed for a E-to-Z photoisomerization with green light, but also allowed reaction with the thiol group to form a covalent bond via nucleophilic aromatic substitution reaction (SNAr) (Hoppmann et al., 2015). This recently developed F-PSCaa 4 has been genetically encoded and was used to trigger conformation changes in CaM and its binding with the nitric oxide synthase (NOS) peptide ligand by visible light. In comparison with UV light, visible light penetrates deeper into tissue and is less toxic, opening new avenues for applying the F-PSCaa system in vivo. Very recently, Deiters and Lin et al. also added new visible light activatable azobenzene amino acids to the genetic code in E. coli and mammalian cells, showing the need and potential of light-sensitive Uaas in addition to conventional optogenetic approaches (John, Ramil, Tian, Cheng, & Lin, 2015; Luo, Samanta, Convertino, Dokholyan, & Deiters, 2018). We found that the synthetase evolved for PSCaas are promiscuous (Hoppmann et al., 2014), possibly allowing other reactive groups in meta positions, such as the keto group in Keto-PSCaa 5, which could undergo keto-oxime ligations.
2.2. Generation of a tRNA/synthetase pair specific for PSCaa incorporation
Expansion of the genetic code with unnatural amino acids (Uaas), a methodology developed almost two decades ago, provides a general platform technology that allows Uaas with various desired properties (e.g. fluorescent, biorthogonal click, bio-reactive etc.) to be site specifically incorporated into proteins in live cells (L. Wang, Brock, Herberich, & Schultz, 2001; L. Wang & Schultz, 2005). The method generates an orthogonal tRNA/synthetase pair specifically for the target Uaa and incorporates the Uaa in response to a unique codon (such as the amber stop codon, UAG) using protein translational machinery. The directed evolution of a synthetase to recognize the Uaa only and none of the endogenous amino acids is a tedious procedure, which involves the generation of a mutant synthetase library (often >109 mutants) followed by selection or screening to identify Uaa-specific synthetase mutants (Santoro, Wang, Herberich, King, & Schultz, 2002; L. Wang & Schultz, 2005). As PSCaas are larger in structure than most genetically encoded Uaas, we prepared the mutant synthetase library by mutating as many residues as possible. Based on rational design and modeling of PSCaa 2 in our previously published X-ray crystal structure of the MmOmeRS, a mutant pyrrolysyl-tRNA synthetase (PylRS) evolved to be specific for Uaa O-methyl-L-tyrosine (Ome), we prepared a mutant PylRS library that allowed us to screen for an appropriate synthetase (Takimoto, Dellas, Noel, & Wang, 2011).
The synthetase obtained from the positive selection showed high orthogonality, i.e. the synthetase does not incorporate endogenous amino acids, and only PSCaas 2-4 are recognized. To determine the efficiency of PSCaa incorporation by the isolated clones, we performed a second quick screen using a mutant GFP gene containing a TAG codon at position 182 and expressed the gene in E. coli in 2xYT medium together with the obtained tRNAPyl/PSCaaRS pairs. PSCaa-dependent GFP expression was observed for 12 clones. The clone that showed the highest selectivity for the incorporation of PSCaa over other amino acids was chosen and its encoded synthetase named MmPSCaaRS. MmPSCaaRS carries the amino acid substitutions A302T, L309S, N346V, C348G.
2.3. Incorporation of F-PSCaa into proteins in E. coli
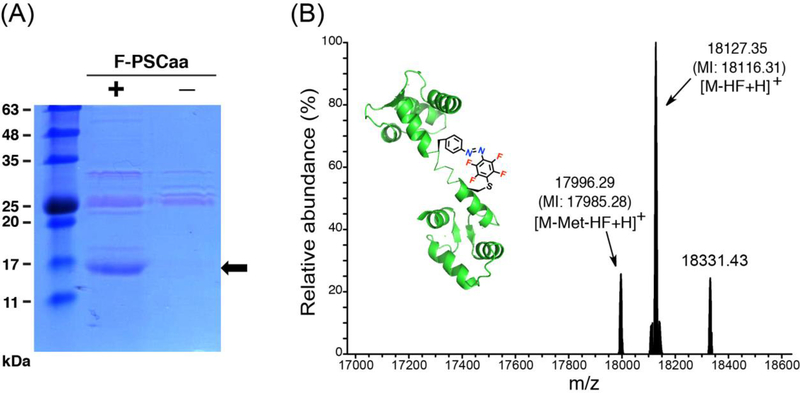
A common way to validate the efficiency and fidelity of an evolved synthetase is to incorporate the Uaa in a model protein of small molecular weight, for which high-resolution mass spectrum of the intact protein can be reliably measured. Model proteins such as Z protein, myoglobin, calmodulin, and ubiquitin have been used for such purposes. To investigate the performance of MmPSCaaRS in incorporating F-PSCaa 3, we expressed in E. coli BL21 cells a gene for calmodulin (CaM76TAGHis6, encoding an amber TAG codon at position 76 and a C-terminal Hisx6 tag) together with the tRNAPyl/MmPSCaaRS. The full-length calmodulin was obtained in good yield (ca. 1.0 mg mL−1) in the presence of 1 mM of F-PSCaa while in the absence of PSCaa no protein expression was detected (Fig. 2A). Electrospray ionization Fourier transform ion trap mass spectrometry (ESI-FTMS) confirmed the incorporation of one F-PSCaa into calmodulin and the formation of the photocontrollable bridge. Upon SNAr reaction of the para-fluorine with the cysteine thiol, HF is released as the leaving group. The corresponding mass for M-HF has been measured (Fig. 2B), and no natural amino acid incorporation at the amber codon position was detected. In addition, no hydrazo myoglobin resulting from the reduction of the azo bond was detected, indicating that the F-PSCaa incorporated into calmodulin remains stable under the reducing conditions of the E. coli cytosol.
Figure 2.
Incorporation of F-PSCaa 4 into CaM forming an azobenzene bridge with cysteine in situ. (A) SDS-PAGE stained with Coomassie shows that tRNAPyl/MmPSCaaRS specifically incorporated F-PSCaa 4 into CaM in E. coli. The arrow indicates the position of the full-length CaM protein. (B) High-resolution ESI-FTMS analysis of intact CaM expressed in the presence of tRNAPyl/MmPSCaaRS supplemented with 1 mM of F-PSCaa 4. Average and monoisotopic (indicated by MI) masses are labeled. The SNAr cross-linking reaction of 4 with cysteine results in the loss of HF. CaM containing the covalent azobenzene bridge: [M − HF + H]+, expected 18116.33 Da, measured 18116.31 Da; [M-Met − HF + H]+, expected 17985.31 Da, measured 17985.28 Da. The peak at 18331.43 did not correspond to CaM with F-PSCaa modified by any known potential intracellular reactants such as Cys, GSH, or imidazole. Adapted with permission from J. Am. Chem. Soc. 2015, 137, 11218–11221. Copyright (2015) American Chemical Society.
3. Expression and photoregulation of PSCaa-incorporated CaM
3.1. Equipment
Fiber optic illuminator and filter (470 nm and 540 nm, EXFO OMNICURE S1000)
Shaker/Incubator
UV-visible spectrophotometer
Analytical balance
Sonicator
Centrifuge
Nanodrop (Thermo Scientific)
CD spectrophotometer (JASCO J-810)
Mass spectrophotometer
FluoroLog spectrophotometer (HORIBA)
3.2. Chemicals
2xYT media (Sigma)
Chloramphenicol (Sigma)
Ni-NTA agarose (Qiagen)
Kanamycin (Thermo Fisher)
F-PSCaa (the synthesis of F-PSCaa is decribed in (Hoppmann et al., 2015))
Tetracycline (Sigma)
Dimethyl sulfoxide (DMSO) (Sigma)
NaOH (0.1 N)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Invitrogen)
Tris-HCl buffer (pH 8) (Sigma)
NaCl (Sigma)
Imidazole (Sigma)
Tween 20 (Sigma)
Glycerol (Sigma)
Lysozyme (Sigma)
Methanol (Sigma)
Dulbecco’s PBS (Sigma)
4. Protocol
4.1. Expression of photoswitchable calmodulin (CaM)
4.1.1 Prepare 2xYT medium and LB-agar plates supplemented with 30 μg mL−1 chloramphenicol and 50 μg mL−1 kanamycin.
4.1.2 Double transform 100 μL electrocompetent E. coli BL21 cells with 1 μL pTak-CaM76TAG plasmid and 1 μL pBK-MmPSCaaRS plasmid (plasmid concentration ~20 ng μL−1) using an electroporator (Eppendorf, EporatorA®) and add 1 mL of prewarmed (37 °C) SOC solution to the bacteria.
4.1.3 Incubate for 60 min at 37 °C in a shaker.
4.1.4 Streak SOC-bacteria solution on a dry LB agar plate (containing chloramphenicol and kanamycin) and place the plate upside down overnight in an incubator at 37 °C.
4.1.5 To make a starter culture, pick one colony from the LB agar plate and grow it overnight in 5 mL 2xYT supplemented with 30 μg mL−1 chloramphenicol and 50 μg mL−1 kanamycin at 37 °C.
4.1.6 This starter culture is used to inoculate 100 mL of 2xYT containing 30 μg mL−1 chloramphenicol and 50 μg mL−1 kanamycin at 37 °C. When OD600 reached 0.5, 1 mM of the unnatural amino acid F-PSCaa was added. Tip: Dissolve the F-PSCaa in DMSO at a concentration of 100 mM in 1 mL and add to 100 mL culture. Alternatively, 1 M NaOH can be used to dissolve PSCaas, but avoid long term storage in solution. After shaking for 5 min, cells were induced for protein expression by adding 0.5 μM IPTG.
4.1.7 After 16 h, cells were pelleted by centrifugation at 10,000 g for 10 min, lysed and sonicated in 5 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole pH 8.0, 1% (v/v) Tween 20, 10% (v/v) glycerol and 0.5 mg mL−1 lysozyme). Lysed cells were centrifuged for 20 min at 14,000 g, and clarified supernatant was incubated with rocking with 0.1 mL Ni2+-NTA agarose resin for 30 min and then passed through the column (Poly-Prep® Chromatography Columns, BioRad). The column was washed with 10 column volumes of wash buffer (lysis buffer without Tween 20 and lysozyme). Protein was eluted with 400 μL of elution buffer (wash buffer containing 250 mM imidazole, pH 8.0). The sample was concentrated to a volume of 50 μL using a Microcon Ultracel YM-10 spin column S2 (Millipore) yielding 1.0 mg mL−1 of CaM as determined by absorbance measurement on nanodrop.
4.1.8 For SDS-PAGE, samples were normalized for constant cell numbers for each lane. Mass spectrometry was performed by Jadebio, Inc. (San Diego).
4.2. Photoswitching of CaM
4.2.1 For E-to-Z photoisomerization, illuminate the solution of CaM (5 μM in dPBS, pH 7.5) bearing the photocontrollable bridge with green light using a fiber optic illuminator with a wavelength filter at λ = 540 nm (5.5 mW cm−2, measured with an EXFO R2000 Radiometer) at r.t. until reaching the photostationary state (pss). The photostationary state is reached when the absorbance spectrum of the irradiated sample remains constant after further irradiation. For CaM at 5 μM concentration pss was reached after 5 min. However, more concentrated solutions (millimolar to molar range) need to be irradiated for longer times.
4.2.2 For Z-to-E isomerization illuminate the solution of CaM bearing the photocontrollable bridge with blue light at λ = 405 or 470 nm (3.9 mW cm−2) for 5 min using a dual fiber optic illuminator and appropriate wavelength filter (405 nm or 470 nm).
4.2.3 Because the E and Z form of the protein do not elute as separate peaks on an HPLC, the isomeric ratio in the pss was estimated based on intensity of absorbances in the UV-visible spectra of the CaM. The absorbance at 337 nm was used to estimate approximately the percentage of E form of CaM, using the WT CaM as the control. The results show that there are E/Z = 31/69 in the 540 nm photoequilibrium (the pss of the Z state) and there are E/Z = 67/33 in the 470 nm photoequilibrium (the pss of the E state).
5. Detecting the photoisomerization of a protein (e.g., CaM)
To follow the photoisomerization of the azobenzene bridge within a protein and see how it influences the protein structure, UV-visible and CD spectra of the ground state (E form) and of the photostationary state (pss) after irradiation were recorded. The change in absorbance intensities of the UV-visible spectra confirms that the azobenzene within the protein was photoactivated and that the isomerization occurred (Fig. 3A). However, UV-visible spectra do not tell us anything about the consequences of the photoisomerization of the azobenzene bridge to the protein structure. To determine whether the protein structure was altered upon photoisomerization of the photocontrollable bridge, CD spectra were recorded (Fig. 3B).
Figure 3.
Azo bridge built on CaM controls CaM conformation and binding. (A) UV−vis spectra of CaM bearing the azobenzene bridge formed by F-PSCaa (incorporated at site 76) reacting with Cys83. Green light illumination (λ = 540 nm, 5 min) resulted in E-to-Z photoisomerization of the built-in azo bridge (black line, before illumination; red line, after green light illumination). Subsequent blue light illumination (λ = 470 nm, 5 min) induced Z-to-E photoisomerization (blue line). (B) CD spectra of the E form (black), the Z pss (red) and the E pss (blue) of CaM bearing the azo bridge in PBS buffer (protein conc. 5 μM). (C) Binding of NOS-I peptide to the E state of CaM/Ca2+ studied by fluorescence. Upon addition of NOS-I to the E form (black line), the fluorescence intensity significantly increased (green line) indicative of peptide binding. (D) CaM/Ca2+ in the E state (black line) was illuminated with green light generating the Z pss state (red line). Subsequent addition of NOS-I peptide to the Z pss state (green line) showed less increase in intensity in comparison to the E state (green line, Fig. 3D). The data obtained suggest that the remaining E form present in the Z pss state binds to the peptide resulting in the slight increase of the intensity. Adapted with permission from J. Am. Chem. Soc. 2015, 137, 11218–11221. Copyright (2015) American Chemical Society.
CD spectroscopy can yield quick information about the global structural change of the protein, but it lacks information about local changes within the protein or a protein domain. To investigate whether the F-PSCaa system allows conformational changes in a protein to be induced by light, we used the well characterized calmodulin as a model protein due to its overall helical conformation. We expressed a CaM gene with a stop codon at position 76 and a cysteine at position 83. The cysteine at position 83 will react with the fluorine in the para position of F-PSCaa creating an azobenzene bridge over an i+7 helical spacing. To determine whether the azobenzene bridge built onto CaM could be directed by visible light, we detected the isomerization process of the purified bridged CaM using UV/vis spectroscopy in PBS buffer.
In the ground state (E form) of the photocontrollable bridge, CaM showed a strong absorbance at 336 nm and a weak band at 445 nm, representing π−π* and n−π* transitions, respectively (black line, Fig. 3A). After green light illumination at λ = 540 nm, the absorbance at 336 nm decreased resulting from the E-to-Z photoisomerization of the azobenzene unit (red line, Fig. 3A). Subsequent illumination with blue light resulted in Z-to-E photoisomerization (blue line, Fig. 3A) yielding the pss of the E state.
To follow how photoisomerization of the azobenzene bridge influences the conformation of the CaM, CD spectra were recorded. The spectra show the typical shape of an α-helical conformation with a band at 222 and 208 nm, respectively (Fig. 3B). Upon E-to-Z photosiomerization the intensity of the n−π* transition band at 208 nm decreased (red line, Fig. 3B), suggesting a conformational change of the CaM protein. Notably, the conformation of the ground E state was restored by Z-to-E photoisomerization using blue light subsequently (blue line, Fig. 3B). The CD signal change detected upon photomodulation has magnitude comparable to that reported for wild-type CaM before and after Ca2+ binding, indicating that photomodulation by the azo bridge resulted in significant conformational change of CaM. Other methods like FTIR, NMR or crystallographic methods can provide more detail about the conformational change, which can be used depending on instrument availability and protein of interest.
To test whether the structural change induced by the embedded photoswitchable bridge allows photomodulation of ligand binding, we measured the binding of CaM to the CaM binding domain of the neuronal nitric oxide synthase (NOS-I). CaM binds the NOS-I peptide by wrapping around the ligand. Those binding events can be easily monitored by fluorescence, so we labeled the azo bridge-containing CaM with dansylchloride in the presence of Ca2+ and recorded dansyl fluorescence upon binding of the NOS-I peptide. After addition of NOS-I peptide (10 μM) to the E form of the bridged CaM/Ca2+ (0.1 μM), the fluorescence intensity significantly increased by 215%. Simultaneously, the emission peak slightly shifted to shorter wavelength (from 517 to 492 nm) as a result of NOS-I peptide binding to CaM (green line, Fig. 3C). In contrast, the change of fluorescence intensity upon addition of NOS-I to the Z pss of CaM/Ca2+ is significantly lower (by 50%, green line, Fig. 3D, the remaining E form present in the Z pss state binds to the peptide resulting in the slight increase of the intensity). This decrease indicates that the CaM after photoswitching to Z form binds less peptide, possibly a result of the conformational change induced by the photoswitch. Illumination of the CaM/Ca2+ in the absence of peptide neither influenced the emission wavelength nor intensity (black and red line, Fig. 3D).
6. SUMMARY
Photopharmacology and optochemical genetics have been reshaping the way in how ion channels and membrane receptors can be modulated by light using chemical tethering of photoswitchable azobenzene-coupled ligands (Hüll, Morstein, & Trauner, 2018, Volgraf et al., 2006, Berlin et al., 2016). Nonetheless, conjugation of the azobenzene ligand is limited mainly to extracellular sites. In contrast, site specific incorporation of Uaa via the expansion of the genetic code allows introduction of the azobenzene switch at any site, even transmembrane regions. Very recently, PSCaa has been incorporated into a set of glutamate receptors (NMDA) in mammalian cells (Klippenstein, Hoppmann, Ye, Wang, & Paoletti, 2017), and multiple sites within the multi-domain receptor were identified showing robust photomodulation. The photoisomerization of PSCaa at the GluN1 clamshell hinge was sufficient for control of glycine sensitivity and activation efficacy; in the pore domain, flipping of a M3 residue within a conserved transmembrane cavity impacts both gating and permeation properties. This study demonstrated detection of molecular rearrangements in real-time due to the reversible light-switching of single amino acid side-chains, adding a dynamic dimension to protein site-directed mutagenesis.
Photocontrollable bridge formation via PSCaa should not be limited to reacting with Cys. Recently, various new latent bioreactive Uaas have been designed and genetically encoded to form covalent bonds with Lys, His, and Tyr on proteins inside live cells via proximity-enabled reactivity(L. Wang, 2017b; N. Wang et al., 2018). These functionalities and reactions can be transferred for building the photocontrollable bridge, in principle. In addition to E. coli, yeast, and mammalian cells, Uaa incorporation via genetic code expansion has been expanded to primary neurons, stem cells, C. elegans, flies, zebrafish, and mice (L. Wang, 2017a), potentially allowing photomodulation of proteins in various cell types and model organisms.
In short, using Uaa technology to generate photoswitchable units on proteins affords a new avenue for optical control of proteins with high specificity and generality. We expect that this method will find broad applications in the emerging field of optobiology for its minimal interference, site-specificity, and genetic encodability.
ACKNOWLEDGMENT
This work was supported by NIH RF1MH114079.
REFERENCES
- Beharry AA, & Woolley GA (2011). Azobenzene photoswitches for biomolecules. Chemical Society Reviews, 40(8), 4422–4437. [DOI] [PubMed] [Google Scholar]
- Berlin S, Szobota S, Reiner A, Carroll EC, Kienzler MA, Guyon A, Xiao T, Trauner D, & Isacoff EY (2016). A family of photoswitchable NMDA receptors. eLife, 5, e12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JG, & Boyden ES (2011). Optogenetic tools for analyzing the neural circuits of behavior. Trends in cognitive sciences, 15(12), 592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose M, Groff D, Xie J, Brustad E, & Schultz PG (2006). The Incorporation of a Photoisomerizable Amino Acid into Proteins in E. coli. Journal of the American Chemical Society, 128(2), 388–389. [DOI] [PubMed] [Google Scholar]
- Brieke C, Rohrbach F, Gottschalk A, Mayer G, & Heckel A (2012). Light-Controlled Tools. 51(34), 8446–8476. [DOI] [PubMed] [Google Scholar]
- Courtney T, & Deiters A (2018). Recent advances in the optical control of protein function through genetic code expansion. Current Opinion in Chemical Biology, 46, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A, Groff D, Ryu Y, Xie J, & Schultz PG (2006). A Genetically Encoded Photocaged Tyrosine. 45(17), 2728–2731. [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, & Deisseroth K (2011). The Development and Application of Optogenetics. 34(1), 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C, Kühne R, & Beyermann M (2012). Intramolecular bridges formed by photoswitchable click amino acids. Beilstein Journal of Organic Chemistry, 8, 884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C, Lacey VK, Louie GV, Wei J, Noel JP, & Wang L (2014). Genetically encoding photoswitchable click amino acids in Escherichia coli and mammalian cells. Angewandte Chemie (International ed. in English), 53(15), 3932–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C, Maslennikov I, Choe S, & Wang L (2015). In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. Journal of the American Chemical Society, 137(35), 11218–11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann C, Schmieder P, Heinrich N, & Beyermann M (2011). Photoswitchable Click Amino Acids: Light Control of Conformation and Bioactivity. 12(17), 2555–2559. [DOI] [PubMed] [Google Scholar]
- Hüll K, Morstein J, & Trauner D (2018). In Vivo Photopharmacology. Chemical Reviews, 118(21), 10710–10747. [DOI] [PubMed] [Google Scholar]
- John AA, Ramil CP, Tian Y, Cheng G, & Lin Q (2015). Synthesis and Site-Specific Incorporation of Red-Shifted Azobenzene Amino Acids into Proteins. Organic Letters, 17(24), 6258–6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-Y, Kawaguchi D, Coin I, Xiang Z, O’Leary DDM, Slesinger PA, & Wang L (2013). In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron, 80(2), 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippenstein V, Hoppmann C, Ye S, Wang L, & Paoletti P (2017). Optocontrol of glutamate receptor activity by single side-chain photoisomerization. eLife, 6, e25808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Samanta S, Convertino M, Dokholyan NV, & Deiters A (2018). Reversible and Tunable Photoswitching of Protein Function through Genetic Encoding of Azobenzene Amino Acids in Mammalian Cells. 19(20), 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudasaini A, El-Arab KK, & Zoltowski BD (2015). LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Frontiers in molecular biosciences, 2, 18-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Wang L, Herberich B, King DS, & Schultz PG (2002). An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nature Biotechnology, 20, 1044. [DOI] [PubMed] [Google Scholar]
- Szymański W, Beierle JM, Kistemaker HAV, Velema WA, & Feringa BL (2013). Reversible Photocontrol of Biological Systems by the Incorporation of Molecular Photoswitches. Chemical Reviews, 113(8), 6114–6178. [DOI] [PubMed] [Google Scholar]
- Takimoto JK, Dellas N, Noel JP, & Wang L (2011). Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient in Vivo Incorporation of Structurally Divergent Non-native Amino Acids. ACS Chemical Biology, 6(7), 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, & Trauner D (2006). Allosteric control of an ionotropic glutamate receptor with an optical switch. Nature chemical biology, 2(1), 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L (2017a). Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts. Accounts of Chemical Research, 50(11), 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L (2017b). Genetically encoding new bioreactivity. New Biotechnology, 38, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brock A, Herberich B, & Schultz PG (2001). Expanding the Genetic Code of Escherichia coli. 292(5516), 498–500. [DOI] [PubMed] [Google Scholar]
- Wang L, & Schultz PG (2005). Expanding the Genetic Code. Angewandte Chemie International Edition, 44(1), 34–66. [DOI] [PubMed] [Google Scholar]
- Wang N, Yang B, Fu C, Zhu H, Zheng F, Kobayashi T, … Wang L (2018). Genetically Encoding Fluorosulfate-l-tyrosine To React with Lysine, Histidine, and Tyrosine via SuFEx in Proteins in Vivo. Journal of the American Chemical Society, 140(15), 4995–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zarrine-Afsar A, Al-Abdul-Wahid MS, Prosser RS, Davidson AR, & Woolley GA (2009). Structure-Based Approach to the Photocontrol of Protein Folding. Journal of the American Chemical Society, 131(6), 2283–2289. [DOI] [PubMed] [Google Scholar]