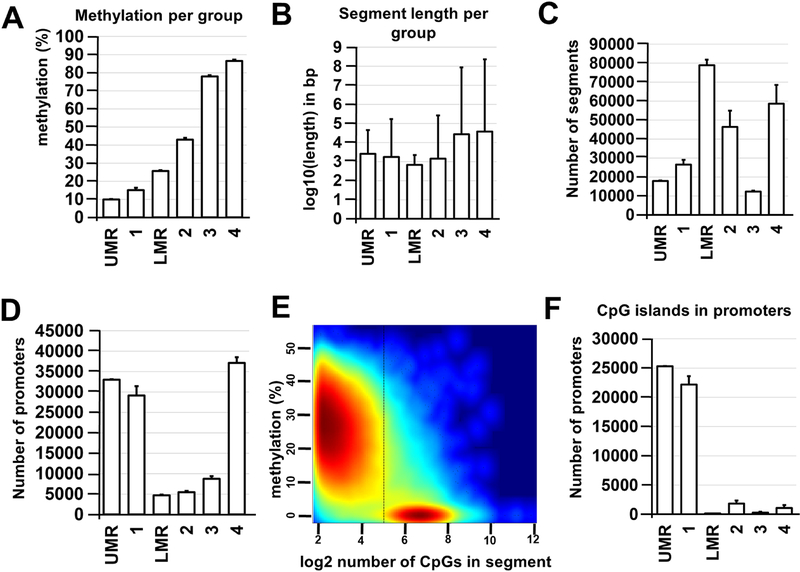
Figure 5.
MethylSeekR and methylKit R Bioconductor packages are robust and comparable methods for identification of Müller glia methylome states. A) Average methylation levels were calculated for each methylation region and segmentation class. B) Average lengths (log10-transformed base pairs; bp) were determined for identified segments and regions. C) Average number of segments vary between segmentation classes and methylation regions. D) The majority of promoters are located in unmethylated (segmentation class 1 and UMR) or highly methylated (segmentation class 4) regions of the Müller glia genome (UMR – unmethylated region; LMR – low-methylated region). E) HMM-based MethylSeekR allowed us to separate CpG-rich UMRs and CpG-poor LMRs. The figure generated by MethylSeekR demonstrates the log2-transformed number of CpGs (hypomethylated regions) versus its average methylation level. F) The CpG islands identified in promoters of Müller glia genomic DNA are mostly unmethylated (segmentation class 1 and UMR).