Fig. 3.
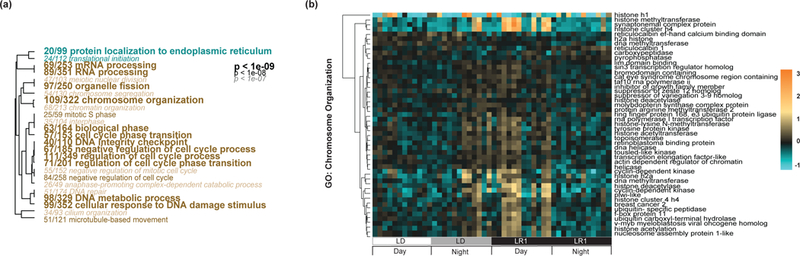
Gene ontology analysis of differently expressed genes after light removal. (A) Significantly up (tan) or down (cyan) -regulated genes related to ‘biological process’ based on a Mann-Whitney U test of the pairwise comparison between the first day following light removal and constant conditions. The font size corresponds to smaller FDR-adjusted p-values, the smallest font is equivalent to a p-value <1e-07, the largest font is equivalent to a p-value <1e-09. Dendrograms represent hierarchical clustering of GO terms based on shared genes in this data set and the ratios in front of each GO term represent the number of genes from that specific GO term in this data set over all genes belonging to that GO term. (B) Clustered heatmap of the top genes (log2 fold change >1.5, DESeq2 p-value <0.001) from the GO term ‘chromosome organization’ (GO:0006325) during light:dark (LD) and one day post light removal (LR1). The color scale is log2 fold change. The experiment key at the bottom identifies the subgroup and the ‘day’ and ‘night’ periods of each 24-hour cycle.
