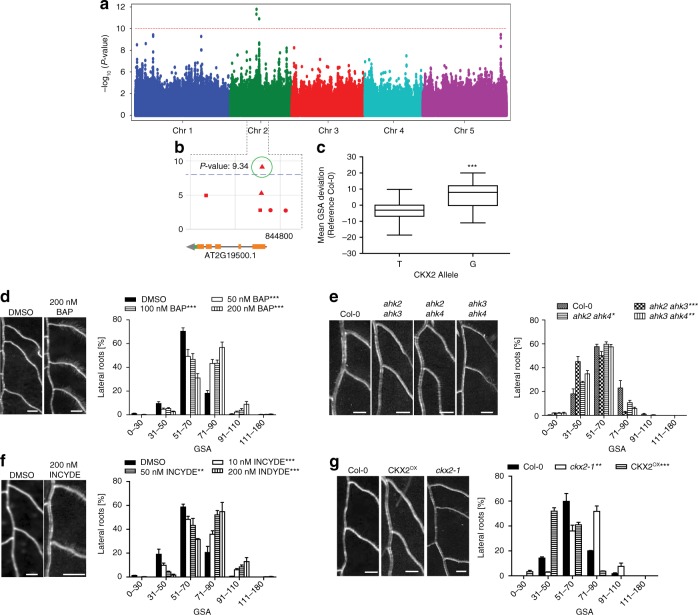
Fig. 2.
Genome-wide association study (GWAS) on gravitropic set point angle (GSA). a Manhattan plot of GWAS results. The dotted horizontal line indicates a significance level of 0.1 after Bonferroni correction for multiple testing. b Magnification of the peak region on chromosome 2. A highly significant SNP was located at position 8,447,233 in the coding region of CKX2. c Mean GSA of T and G alleles of CKX2. Horizontal lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the min and max values. Student’s t-test P-value: ***P < 0.001. d–g Representative images and GSA distributions of untreated and 6-Benzylaminopurin (BAP)-treated Col-0 wild type d, Col-0 wild type, ahk2 ahk3, ahk2 ahk4 and ahk3 ahk4 e, untreated and INCYDE-treated Col-0 wild type f, Col-0 wild type, ckx2-1 and CKX2OX seedlings g. Kolmogorov–Smirnov test P-values: *P < 0.05, **P < 0.01, ***P < 0.001 (compared to DMSO solvent or Col-0 wild type control). Mean ± SEM, n = 5 plates (16 seedlings with 65–160 LRs per plate). Scale bars, 2 mm. d–g Experiments were repeated at least three times