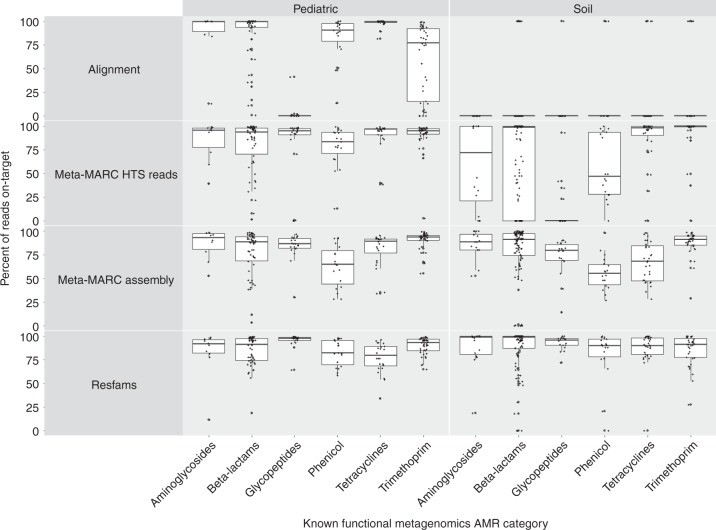
Fig. 3.
AMR class abundance for the shotgun metagenomic dataset of Noyes et al.32. Alignment (green), Meta-MARC HTS (orange), Meta-MARC Assembly (blue) and Resfams (purple) are compared. Meta-MARC Assembly identified the greatest number of reads in 11 out of 13 resistance classes. For the aminoglycosides and multi-drug resistance genes, Resfams identified a greater number of reads with Meta-MARC containing the second largest number of identifications in those classes. Overall, Meta-MARC Assembly detected 42-fold more reads than Alignment, and 1.5-fold more reads than Resfams