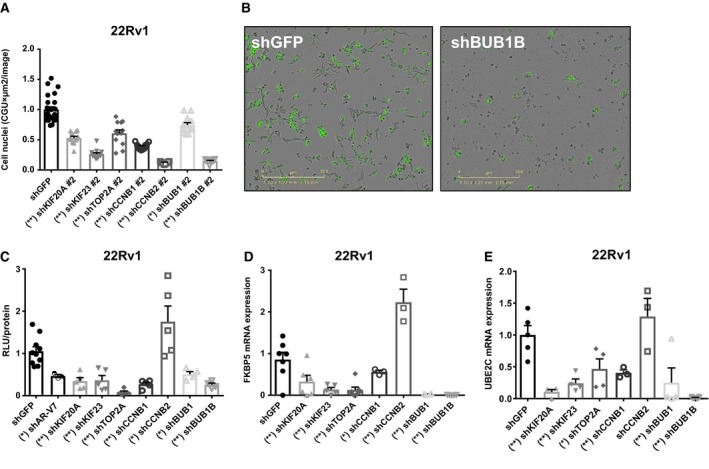
Figure 3. Depletion of the expression of members of the seven‐gene set reduces CRPC cell proliferation and AR ligand‐independent transcriptional activity.
-
ACell proliferation was examined in the CRPC cell line 22Rv1 following individual depletion of mRNAs for the seven genes or shGFP controls, using shRNA against the coding region for each gene (shRNA #2). Cell number was measured using a non‐perturbing nuclear restricted dye and quantified after 72 h using Incucyte Zoom System. Data shown are mean ± s.e.m. of eight to 12 replicates normalized to their shGFP control. Kruskal–Wallis test (P‐value < 0.0001, two‐tailed) and Dunn's multiple comparisons test were performed. *P‐value < 0.05, **P‐value < 0.001.
-
BRepresentative images of 22Rv1 stably depleted of BUB1B or control (shGFP) are shown.
-
C22Rv1 stably depleted of each of the seven genes were transfected with a dual‐plasmid luciferase reporter system which quantifies AR activity and basal transcription. The assay was conducted in 2% CSS to measure AR ligand‐independent transcriptional activity. Data represent two independent experiments performed in triplicate, showing the mean ± s.e.m., and normalized to their shGFP controls. Kruskal–Wallis test (P‐value < 0.0001, two‐tailed) and Dunn's multiple comparisons test were performed. *P‐value < 0.05, **P‐value < 0.001.
-
D, EThe expression of FKBP5 and UBE2C determined by RT–qPCR analysis and normalized to GAPDH mRNA levels was examined in 22Rv1 cells stably expressing shRNA for each of the seven genes. Cells were cultured in 2% CSS. Data represent two independent experiments performed in duplicate or triplicate, showing the mean ± s.e.m., and normalized to their shGFP controls. Kruskal–Wallis test (P‐value < 0.0001, two‐tailed) and Dunn's multiple comparisons test were performed. *P‐value < 0.05, **P‐value < 0.001.
Source data are available online for this figure.
