FIGURE 1.
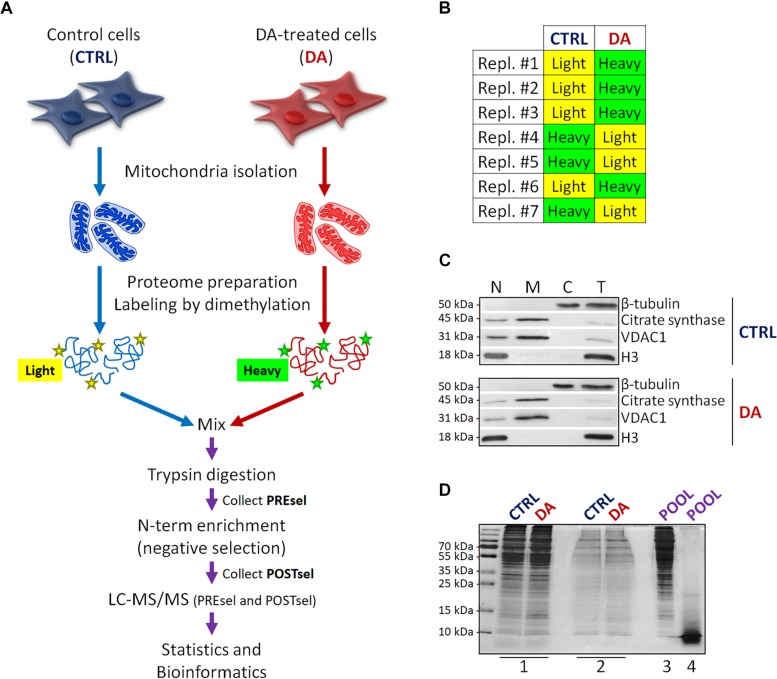
Mitochondrial dimethylation-TAILS. (A) Outline of the experimental workflow designed to investigate mitochondrial proteases and their substrates in a cellular model of altered DA homeostasis. After dopamine treatment, mitochondria were isolated from both control (CTRL) and DA-treated (DA) SH-SY5Y cells, isotopic labeling of primary amines was performed by either light or heavy dimethylation, samples were mixed, proteins were digested by trypsin and N-terminal peptides were enriched by negative selection, prior to perform the LC-MS/MS analysis. Both pre- (PREsel) and post- (POSTsel) negative selection samples have been collected and analyzed. (B) Table representing the labeling strategy for the seven biological replicates. (C) Representative WB analysis of fractions collected after mitochondria isolation. N, nuclei; M, mitochondria; C, cytoplasm; T, total extract. (D) TAILS quality control SDS-PAGE representing one in seven biological replicates. 1: mitochondrial proteins after lysis and first precipitation; 2: after isotopic labeling; 3: after pooling and second precipitation; 4: after trypsin digestion. See “Materials and Methods” section for further details.