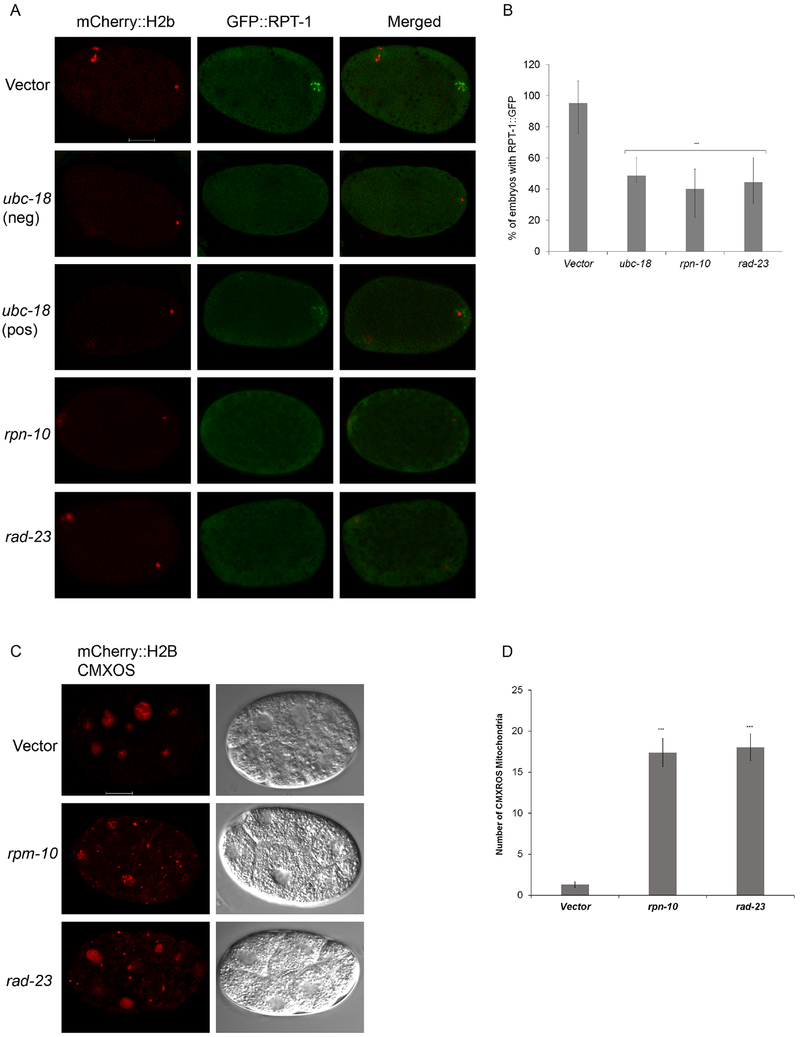
Figure 4. Proteasome recruitment requires ubc-18, rpn-10, and rad-23.
(A) Transgenic worms expressing mCherry::H2B and RPT-1::GFP fusion were treated with either vector, ubc-18, rad-23, or rpn-10 RNAi. In vector treated meiosis I embryos, RPT-1::GFP localizes to the region around the sperm DNA. RNAi of ubc-18, rpn-10, or rad-23 reduces the amount of RPT-1::GFP localizing around the sperm DNA. One embryo that was scored as positive for GFP::RPT-1 puncta after ubc-18(RNAi) is also shown (third row). Even though the puncta are present, the level of GFP at those puncta is lower than that seen in the control. (B) In about half of the RNAi treated embryos early proteasome recruitment was absent. 30 Meiosis I embryos per treatment were observed. Statistical significance was calculated by a Fisher’s Exact test: ***p < 0.001. Error bars represent 95% confidence intervals. (C) Paternal mitochondria were tracked in 8-12 cell embryos using CMXROS labeled males. Embryos from rpn-10 and rad-23 knockdowns showed larger numbers of paternal mitochondria persisting in 2-12 cell embryos when compared to the control. Both paternal mitochondria (CMXROS) and maternal DNA (mCherry::H2B) appear red in these images. (D) Quantification of mitochondria from 8-10 cell embryos. 12 embryos per trial with a total of 2 trials. Data in the graph represent the mean ± s.e.m. Significant differences between embryos was determined by unpaired t test. ***p<000.1.