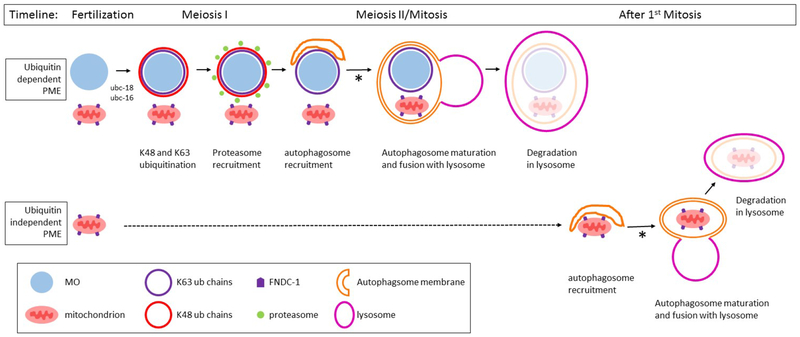
Figure 7. Model for the role of ubiquitination in the elimination of paternal mitochondria in C. elegans.
The targeting of paternal mitochondria for elimination (PME) after fertilization involves at least two separate pathways: ubiquitin dependent and ubiquitin independent pathways. In the ubiquitin dependent pathway, the membraneous organelles (MO) are quickly decorated with K48 (red) and K63 (blue) polyubiquitin chains. The MOs (light blue) and paternal mitochondria (pink) remain clustered together until the beginning of mitosis. The K48 chains recruit proteasomes (green) to the MO surface. K48 chains and proteasomes remain associated with the MOs for only a few minutes until the completion of meiosis I. After meiosis I, the autophagosome membranes (orange) are recruited to MOs and mitochondria. These autophagosomes presumably fuse with lysosomes and lead to the degradation of the enveloped MOs and mitochondria. Paternal mitochondria that escape the process described above can be degraded by a ubiquitin independent mechanism. The FNDC-1 protein (purple pentagon) localizes to the surface of paternal mitochondria, recruiting autophagosomal membranes via its LIR domain. These autophagsomes presumably fuse with lysosomes and lead to the degradation of the encased mitochondria. The process of autophagosomal maturation and lysosome fusion in both pathways requires the proteasomal associated ubiquitin recognition proteins RAD-23 and RPN-10 (denoted with an asterisk).