Figure 2. Apelin deletion delays VEGF‐induced blood vessel sprouting.
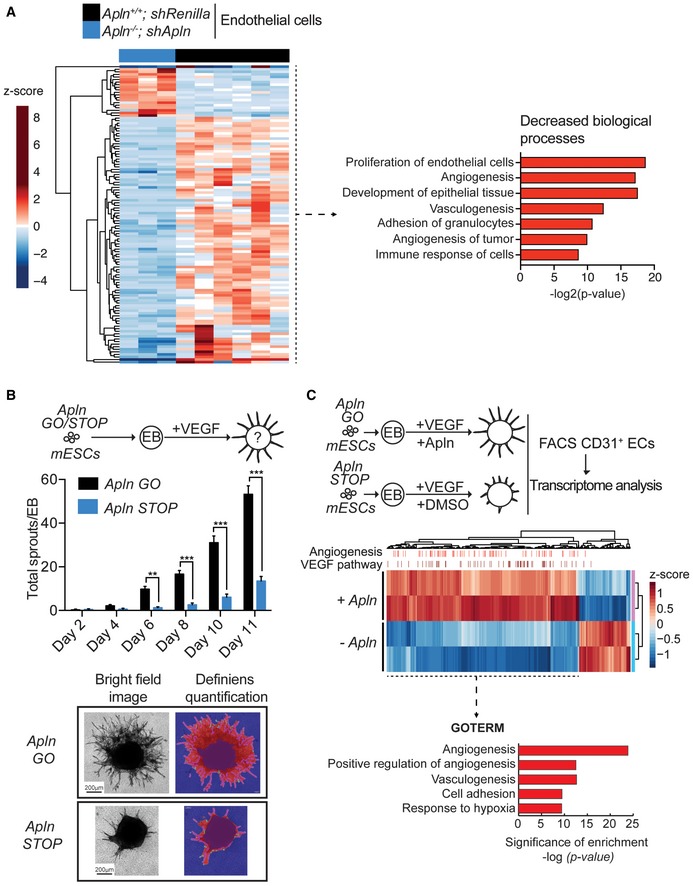
- Left—Heatmap of RNA‐Seq transcriptome analysis of CD31+/CD105+ endothelial cells sorted from tumors established by E0771 shRenilla (n = 6) or shApln (n = 3) cells orthotopically injected into C57BL/6J Apln +/+ or Apln −/− mice, respectively. Tumors were harvested day 25 post‐injection and genes displayed are significantly deregulated at the adjusted P‐value cutoff of 0.05. Right—ingenuity pathway analysis for biological processes predicted to be decreased downstream of the differentially expressed genes.
- Quantification of vessel sprouts (mean values ± SEM) upon VEGF treatment (30 ng/ml) of embryoid bodies (EB) derived from murine ES cells (mESCs) with sense integrations in the Apelin gene (Apln STOP) of the splice acceptor described in Appendix Fig S1B or Cre‐reverted antisense (Apln GO) sister cells. Apln GO (Day 2 n = 6; Day 4 n = 40, Day 6 n = 54, Day 8 n = 32, Day 10 n = 35, Day 11 n = 31), Apln STOP (Day 2 n = 4; Day 4 n = 15, Day 6 n = 31, Day 8 n = 28, Day 10 n = 32, Day 11 n = 37); **P < 0.01, ***P < 0.001, two‐way ANOVA. Representative brightfield images and automated analysis of vessel sprouts by Definiens software are shown in the bottom panels. Scale bars = 200 μm.
- Differentially expressed genes using RNA‐Seq transcriptome analysis of CD31+ endothelial cells (ECs) sorted from sprouting EBs from Apelin STOP cells stimulated with VEGF (30 ng/ml) and DMSO (−Apln) and repaired Apelin GO sister cells stimulated with VEGF and Apelin (1,000 nM; +Apln). VEGF target genes and angiogenesis‐related genes, predicted by ingenuity pathway analysis (IPA) software, are indicated by bars on the upper axis of the heatmap. GO terms were analyzed by DAVID online software.
