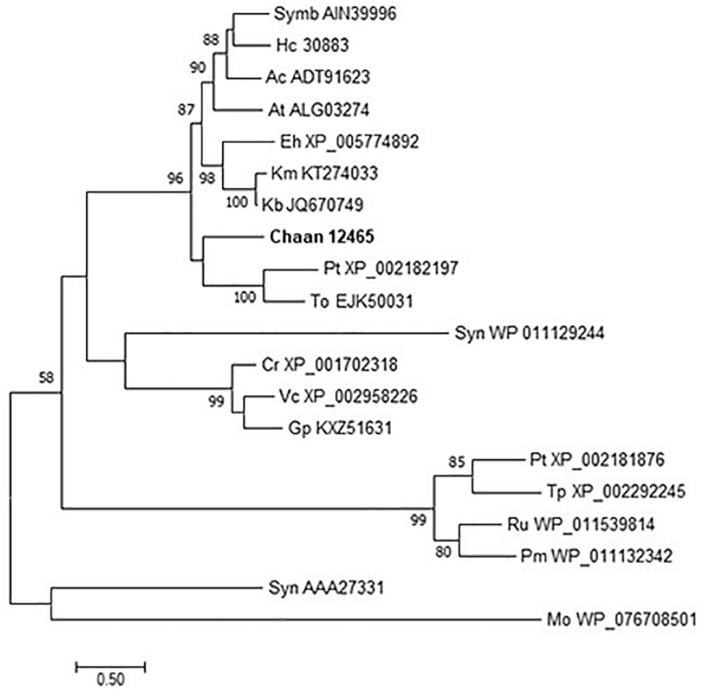
FIGURE 7.
The maximum-likelihood phylogenetic tree of alkaline phosphatase genes. The tree was inferred using the WAG+G model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates. Each gene is labeled with the abbreviated scientific name and accession ID. Abbreviations of species names are as follows: (Raphidophyceae) Chaan, Chattonella antiqua; (Bacillariophyceae) Pt, Phaeodactylum tricornutum; To, Thalassiosira oceanica; Tp, Thalassiosira pseudonana; (Chlorophyceae) Cr, Chlamydomonas reinhardtii; Gp, Gonium pectoral; Vc, Volvox carteri; (Dinophyceae) At, Alexandrium tamarense; Ac, Amphidinium carterae; Hc, Heterocapsa circularisquama; Kb, Karenia brevis; Km, Karenia mikimotoi; Symb, Symbiodinium sp.; (Prymnesiophyceae) Eh, Emiliania huxleyi; (cyanobacteria) Syn, Synechocystis sp.; Pm, Prochlorococcus marinus; Syn, Synechococcus sp.; (bacteria) Mo, Microbacterium oleivorans; and (proteobacteria) Ru, Ruegeria sp.