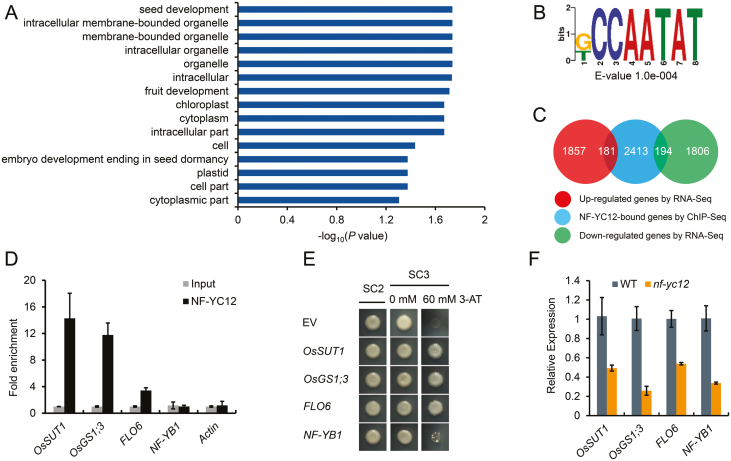
Fig. 7.
Overview of ChIP-seq data and identification of NF-YC12 direct target genes in rice. (A) Enriched gene ontology (GO) terms of the genes bound by NF-YC12 as determined by ChIP-seq analysis. Only GO terms with a corrected P-value <0.05 and including at least five annotated genes were kept. The length of the bars represents the negative logarithm (base 10) of the corrected P-value. (B) Motif analysis of NF-YC12 binding peaks by DREME. The ‘CCAATA’ (CCAAT-box) motif was identified as one of the top five enriched motifs. The E-value is the enrichment P-value multiplied by the number of candidate motifs tested. The enrichment P-value was calculated using Fisher’s exact test for enrichment of the motif in the positive sequences. (C) Venn diagram showing the number of overlapping genes between the NF-YC12-bound gene set (ChIP-seq data) and the NF-YC12-regulated gene set (RNA-seq). (D) ChIP-PCR verification of NF-YC12-bound regions. The data are the mean values (±SD) of fold-enrichment from n=3 technical replicates. (E) The interaction between NF-YC12 and the promoters of target genes as determined by yeast one-hybrid analysis. EV, empty vector; SC2,: SD/–Leu/–Trp; SC3, SD/–Leu/–Trp/–His. (F) qRT-PCR analysis of expression levels of the target genes in nf-yc12 Compared with the wild-type (WT). Ubiquitin was used as the reference gene.