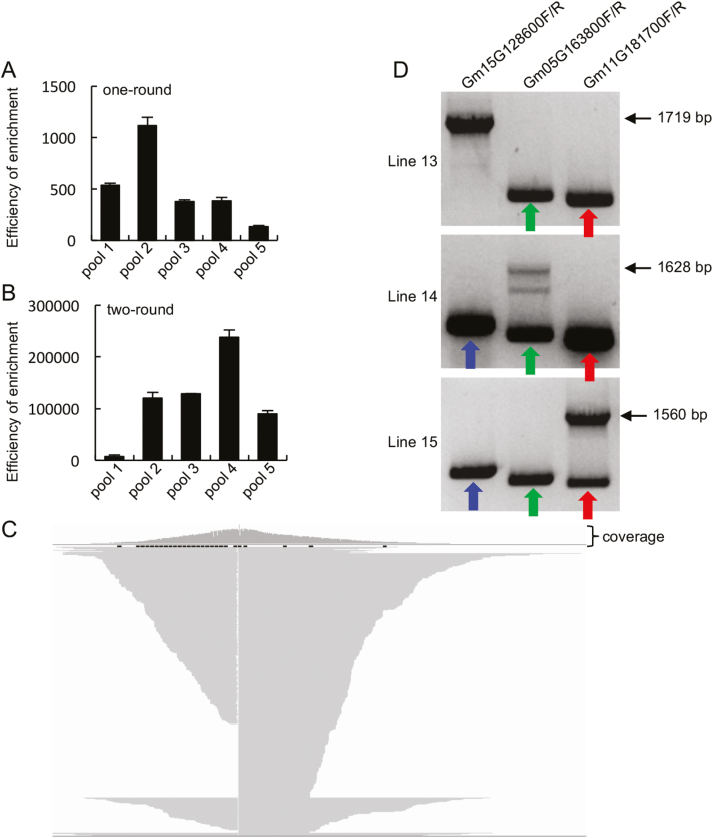
Fig. 4.
Sequencing results after two-round enrichment of the Ds-containing fragments. (A and B) Efficiency of one-round (A) and two-round (B) enrichment of the Ds element-containing fragments; 2% of samples before and after probe enriching were used to perform qPCR. The amount of target fragments was normalized to that of the internal control. (C) Schematic diagram of the flanking sequences of Line 15. Partial sequences of reads are shown. (D) PCR validation of the Ds insertion in Lines 13, 14, and 15. The three individual lines were examined with three pairs of primers each. Each primer pair (labeled above the picture) recognizes a potential insertion position of the Ds element, identified by sequencing. Line 13 containing a Ds insertion in the Glyma15G128600 gene produced a 1719 bp fragment, while Lines 14 and 15 without insertions in this gene generated ~559 bp fragments (indicated with arrows). Line 14 containing a Ds insertion in the Glyma05G163800 gene produced a 1628 bp fragment, while Lines 13 and 15 without insertions in this gene generated 462 bp fragments (indicated with arrows). Line 15 containing a Ds insertion in the Glyma11G181700 gene produced a 1560 bp fragment, while Lines 13 and 14 without insertions in this gene generated 394 bp fragments (indicated with arrows).