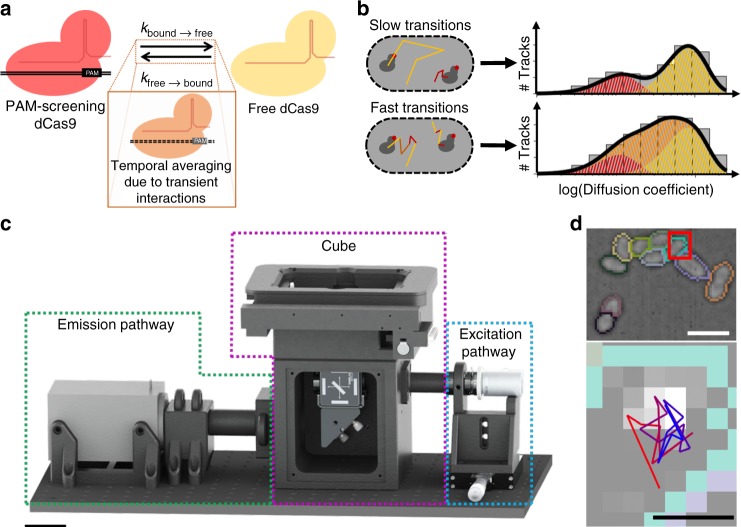
Fig. 1.
Probing cellular dynamics of dCas9 on an open-source microscope using sptPALM. a Simplified expected dynamic behaviour of dCas9 in absence of DNA target sites. The protein can be temporarily bound to DNA (PAM screening), or diffuse freely in cytoplasm, with two kinetic rates governing the dynamics. If the interaction is on a similar timescale as the detection time, a temporal averaging due to transient interactions is expected. b If the dynamic transitions are slow with respect to the camera frame time used in sptPALM, the obtained diffusional data can be fitted with a static model (top), which assumes that every protein is either free (yellow) or DNA-bound (red), but does not interchange. If the dynamic transitions are as fast or faster than the frame time used, Monte-Carlo diffusion distribution analysis (MC-DDA; bottom) can fit the diffusional data. In MC-DDA, dCas9 can interchange between the two states, resulting in a broader distribution. c Render of the open-source miCube super-resolution microscope. The excitation components, main cube, and emission components are indicated in blue, magenta, and green, respectively. Details are provided in the “Methods” section. Scale bar represents 5 cm. d Brightfield images of L. lactis used for computationally obtaining the outline of the cells via watershed (top), and raw single molecule data (bottom; red outline in top is magnified) as obtained on the miCube as part of a typical experiment, overlaid with the determined track where this single molecule belongs to (starting at red, ending at blue). Scale bars represent 2.5 µm (top) or 500 nm (bottom)