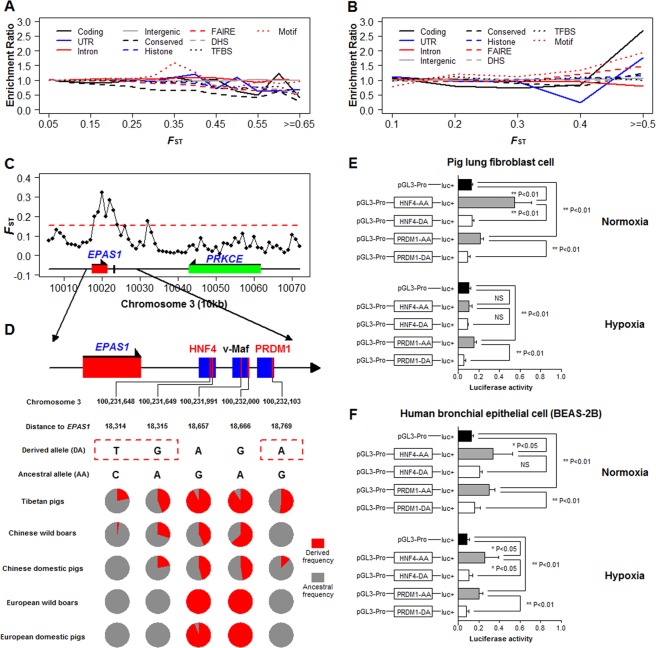
Figure 4.
Enrichment of variants under different divergence levels and transcription activity assay of alleles in predicted motifs near EPAS1. (A) Enrichment pattern of SNPs at whole genome level. SNPs were divided to different bins by FST between Tibetan and lowland pigs. (B) Enrichment pattern of SNPs from selected sweep regions in Tibetan pigs. (C) Selective signals of EPAS1. The black vertical bar in downstream of EPAS1 indicated differentiated SNP positions between Tibetan and lowland pigs. The red dotted line represents top 99% threshold of FST at whole-genome level. (D) Predicted motifs with differentiated SNPs in downstream of EPAS1. (E) Transcription activity assay of different alleles within predicted motifs near EPAS1 in pig lung fibroblast cell. AA names of different pGLS3 vectors means ancestral allele and DA means derived allele. (F) Transcription activity assay of different alleles within predicted motifs near EPAS1 in BEAS-2B cell. The two-tailed t test was used for statistical assessment of transcription activity change.