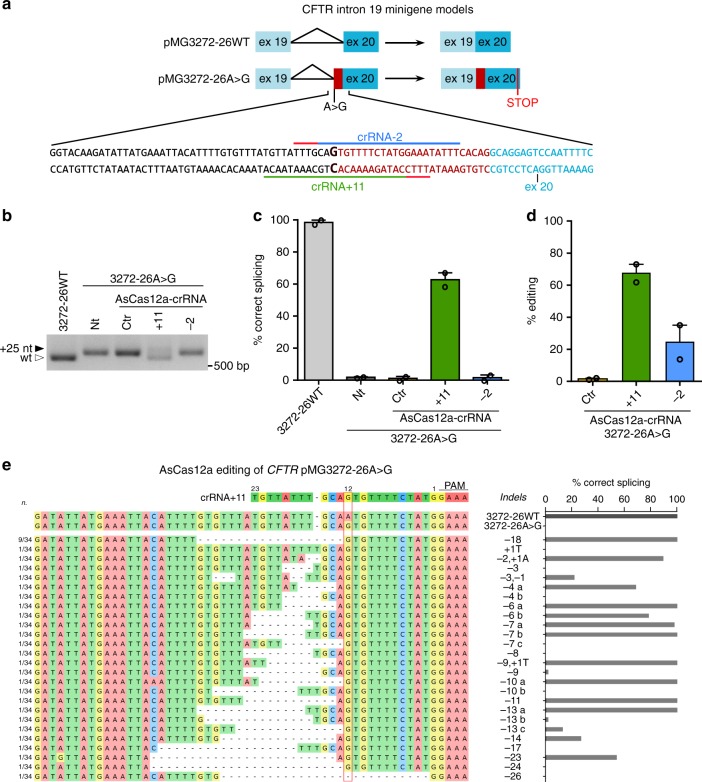
Fig. 1.
Splicing correction in CFTR 3272-26A>G minigene model by AsCas12a editing. a Scheme of the CFTR minigenes containing a sequence (~1.3 kb) corresponding to the CFTR region extending from exon 19 to 20 either wild type (pMG3272-26WT) or 3272-26A>G mutated (pMG3272-26A>G). Exons are shown as boxes, introns as lines; the expected spliced transcripts are represented on the right according to the presence or absence of the 3272-26A>G mutation. The lower panel shows nucleotide sequence and intron–exon boundaries nearby the 3272-26A>G mutation (labeled in bold) and the target AsCas12a-crRNAs position (underlined, with the PAM in red). b Splicing pattern analyzed by RT-PCR in HEK293/pMG3272–26A>G cells following treatments with AsCas12a-crRNA control (Ctr) or specific for the 3272-26A>G mutation (+11 and −2). Black-solid arrow indicates aberrant splicing, white-empty arrow indicates correct splicing. Representative data of n = 2 independent experiments. c Percentages of correct splicing measured by densitometry and d editing efficiency analyzed by TIDE in cells treated as in b. Data are means ± SEM from n = 2 independent experiments. e Indels triggered by AsCas12a-crRNA+11. The 3272-26A>G locus from cells edited by crRNA +11 were amplified, cloned in the minigene backbone and Sanger sequenced (34 different clones, left panel), or analyzed as in b for produced splicing pattern (right panel and Supplementary Fig. 3b). pMG3272-26WT and pMG3272-26A>G were used as references