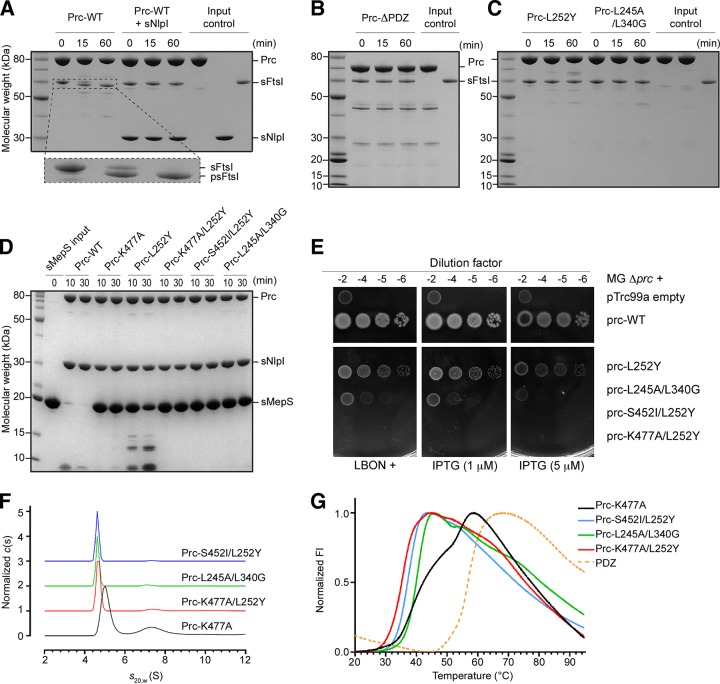
FIG 1.
Biochemical and biophysical properties of Prc and various mutants. (A) SDS-PAGE analysis monitoring the cleavage of FtsI missing the N-terminal transmembrane segment (residues 57 to 588; sFtsI) into the processed form (residues 57 to 577; psFtsI) by wild-type Prc with and without NlpI (expressed without the lipid anchor; sNlpI). The dashed area of the gel is enlarged and shown below to highlight the cleavage reaction. (B) Assays of sFtsI cleavage by Prc with the PDZ domain deleted. (C) Assays of sFtsI cleavage by Prc with mutations in the PDZ ligand-binding pocket (L252Y) and the substrate-sensing residues (L245A and L340G). (D) Assays of degradation of MepS, expressed without the lipid anchor (sMepS), by wild-type Prc and various Prc mutants. (E) Viability assays examining the effects of various Prc mutations on the growth of the Δprc mutant (MG Δprc) on medium of low osmotic strength (LBON). Wild-type prc in the pTRC99a plasmid has a leaky expression sufficient to complement the Δprc mutant even without the inducer IPTG. (F) Sedimentation velocity profiles comparing the molecular sizes of various Prc double mutants and Prc-K477A, which was in the liganded activated state (4). (G) DSF melting curves comparing the melting transitions of the Prc double mutants with liganded Prc-K477A and the isolated PDZ domain. FI, fluorescence intensity.