FIG 5.
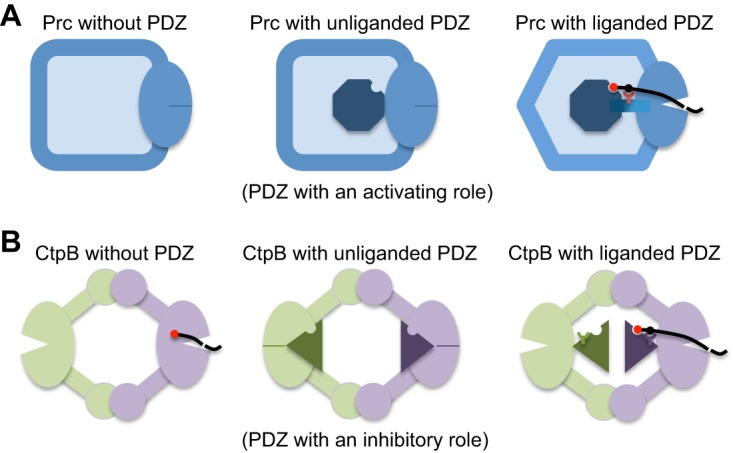
Comparison of the structural mechanisms for the regulation of the protease activity of Prc and CtpB by the PDZ domain. (A) In Prc, the PDZ deletion results in an inactive protease (left), as evidenced by a misaligned protease active site (indicated by a solid dash). In the resting state, the unliganded PDZ domain (the octagon) docks inside the bowl-like scaffold structure of Prc and makes no contact with the protease active site (middle). Upon substrate binding, the hydrophobic sensor (Leu340; indicated by a Y-shaped symbol) engages the bound substrate and triggers structural remodeling to align the protease active site for substrate cleavage (right). (B) In dimeric CtpB, deletion of the PDZ domain yields a constitutively active protease (left). In the inhibited resting state, the protease active site is disrupted by the docked PDZ domain (triangles) (middle). Substrate binding induces repositioning of the PDZ domain, stabilized by the polar sensor (Arg168) from the PDZ domain (right).
