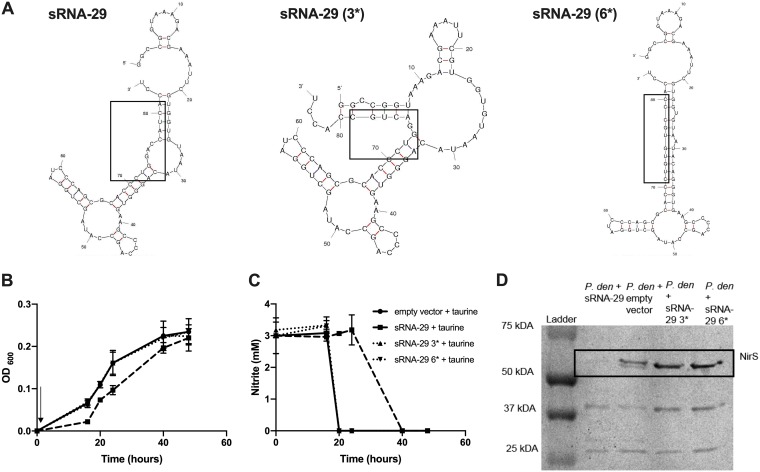
FIG 4.
Mutation of either 3 or 6 nucleotides of sRNA-29 alters the predicted secondary structure, which results in a loss of function under conditions of growth on nitrite. (A) Mutation of 3 (3*) or 6 (6*) nucleotides of sRNA-29 altered the secondary structure as predicted by Mfold. P. denitrificans plus empty pLMB509 vector (solid line) or sRNA-29 or sRNA-29 with 3 (3*) or 6 (6*) nucleotides mutated (various dashed lines and symbols as indicated in legend) was grown under denitrification conditions (3 mM nitrite as electron acceptor) with 10 mM taurine added to induce sRNA-29 overexpression at time zero (indicated with an arrow in panel B). OD600 levels (B) and nitrite consumption levels (C) were measured. Error bars represent SD of results of comparisons between triplicate experiments, and where not visible, the bars were smaller than the symbols. (D) Soluble protein was extracted from cultures at 24 h and subjected to SDS-PAGE and subsequent heme staining.