The discovery of antibiotics to treat bacterial infections has had a dramatic and positive impact on human health. However, shortly after the introduction of a new antibiotic, bacteria often develop resistance. The bacterial cell envelope is essential for cell viability and is the target of many of the most commonly used antibiotics, including β-lactam antibiotics. Resistance to β-lactams is often dependent upon β-lactamases. In B. cereus, B. thuringiensis, and some B. anthracis strains, the expression of some β-lactamases is inducible. This inducible β-lactamase expression is controlled by activation of an alternative σ factor called σP. Here, we show that β-lactam antibiotics induce σP activation by degradation of the anti-σ factor RsiP.
KEYWORDS: cell envelope, extracellular signaling, gene expression, sigma factors, signal transduction, stress response
ABSTRACT
Bacteria can utilize alternative σ factors to regulate sets of genes in response to changes in the environment. The largest and most diverse group of alternative σ factors are the extracytoplasmic function (ECF) σ factors. σP is an ECF σ factor found in Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Previous work showed that σP is induced by ampicillin, a β-lactam antibiotic, and required for resistance to ampicillin. However, it was not known how activation of σP is controlled or what other antibiotics may activate σP. Here, we report that activation of σP is specific to a subset of β-lactams and that σP is required for resistance to these β-lactams. We demonstrate that activation of σP is controlled by the proteolytic destruction of the anti-σ factor RsiP and that degradation of RsiP requires multiple proteases. Upon exposure to β-lactams, the extracellular domain of RsiP is cleaved by an unknown protease, which we predict cleaves at site-1. Following cleavage by the unknown protease, the N terminus of RsiP is further degraded by the site-2 intramembrane protease RasP. Our data indicate that RasP cleavage of RsiP is not the rate-limiting step in σP activation. This proteolytic cascade leads to activation of σP, which induces resistance to β-lactams likely via increased expression of β-lactamases.
IMPORTANCE The discovery of antibiotics to treat bacterial infections has had a dramatic and positive impact on human health. However, shortly after the introduction of a new antibiotic, bacteria often develop resistance. The bacterial cell envelope is essential for cell viability and is the target of many of the most commonly used antibiotics, including β-lactam antibiotics. Resistance to β-lactams is often dependent upon β-lactamases. In B. cereus, B. thuringiensis, and some B. anthracis strains, the expression of some β-lactamases is inducible. This inducible β-lactamase expression is controlled by activation of an alternative σ factor called σP. Here, we show that β-lactam antibiotics induce σP activation by degradation of the anti-σ factor RsiP.
INTRODUCTION
The bacterial cell envelope is essential for cell viability and is the target of many of the most commonly used antibiotics, including β-lactams like penicillins, penems, and cephalosporins. These are broad-spectrum antibiotics that target peptidoglycan (PG) biosynthesis by inhibiting the transpeptidase activity of penicillin-binding proteins (PBPs). This results in decreased and/or altered cross-linking of peptidoglycan, which leads to cell envelope damage and subsequent cell lysis and death (1, 2).
Members of the Bacillus cereus group, including Bacillus thuringiensis and Bacillus cereus and some strains of Bacillus anthracis, are highly resistant to β-lactam antibiotics (3–6). This resistance is due in part to expression of at least two β-lactamases (3, 5). The expression of these β-lactamases is induced by ampicillin and is dependent upon the alternative σ factor σP. σP belongs to the extracytoplasmic function (ECF) family of alternative σ factors (5).
Bacteria often utilize alternative σ factors to regulate subsets of genes required for survival under specific environmental conditions or for stress responses. ECF σ factors are the largest and most diverse group of alternative σ factors and represent the “third pillar” of bacterial signal transduction (7, 8). ECF σ factors belong to the σ70 family, but unlike the “housekeeping” σ factor, σ70, ECF σ factors contain only region 2 and region 4.2 of σ70, which recognize and bind to the −10 and −35 regions of promoter sequences, respectively (8, 9). In addition, unlike σ70, ECF σ factors are generally held inactive by anti-σ factors until bacteria encounter an inducing signal (10, 11). Upon induction, ECF σ factors are released from their cognate anti-σ factors to promote transcription of specific stress response genes.
The ECF σ factors have been subdivided into more than 40 distinct groups, with ECF01 being the best studied (reviewed in references 7, 11, and 12). σP belongs to the ECF01 family, which includes members like σE and σW from Escherichia coli and Bacillus subtilis, respectively. The activities of the ECF01 family are inhibited by their cognate transmembrane anti-σ factors (8, 13). To activate ECF01 σ factors, the anti-σ factors must be destroyed via a proteolytic cascade (14, 15). For example, the E. coli anti-σ factor RseA is degraded in response to outer membrane stress, leading to σE activation (16, 17). DegS, a serine protease, cleaves the anti-σ factor RseA at site-1 (14, 18, 19). After site-1 cleavage, the conserved site-2 protease, RseP, cleaves RseA within the membrane, leading to increased σE activity (14, 20, 21). Similarly, the σW anti-σ factor, RsiW, from B. subtilis is proteolytically degraded by site-1 and site-2 proteases. In the case of RsiW, the site-1 protease is PrsW, a metalloprotease unrelated to DegS. PrsW cleaves RsiW in response to antimicrobial peptides, vancomycin, and pH change (22–24). RsiW is further processed by the conserved site-2 protease RasP, a homolog of RseP (15).
The closely related ECF30 family member σV from B. subtilis is activated by lysozyme (25–29). Activation of σV differs from σE and σW activation in that σV is not controlled by a dedicated site-1 protease but instead utilizes signal peptidases (30, 31). Signal peptidases are essential proteases which are required to cleave substrates secreted from the general secretion or twin arginine secretion systems (32–34). The anti-σ factor RsiV binds to lysozyme, which allows signal peptidase to cleave RsiV at site-1 (30, 31). This allows the site-2 protease RasP to cleave RsiV, leading to σV activation (35).
Previous studies found that σP is induced by ampicillin (Amp) and that its activity is required for resistance to ampicillin (5). The activity of σP is inhibited by the transmembrane anti-σ factor RsiP (5, 6). However, whether σP is activated specifically by ampicillin or more generally by cell wall stress is not known. In B. subtilis, activation of σV is specific to lysozyme (26, 27), while activation of σW, σX, and σM is in response to more general cell envelope stress (9, 36, 37). Here, we show that σP is activated by a specific subset of β-lactams and that this activation occurs via regulated intramembrane proteolysis of the anti-σ factor RsiP.
RESULTS
A subset of β-lactams induces σP activation.
Previously, Koehler and colleagues demonstrated that ampicillin induces expression of the β-lactamase encoded by bla1 (hd73_3490) in a σP-dependent manner in B. thuringiensis and B. cereus (5). Activation of some ECF σ factors is highly specific to an inducing signal, while others are activated by more general cell envelope stress. Thus, we sought to determine the specificity of σP activation using B. thuringiensis as a model system.
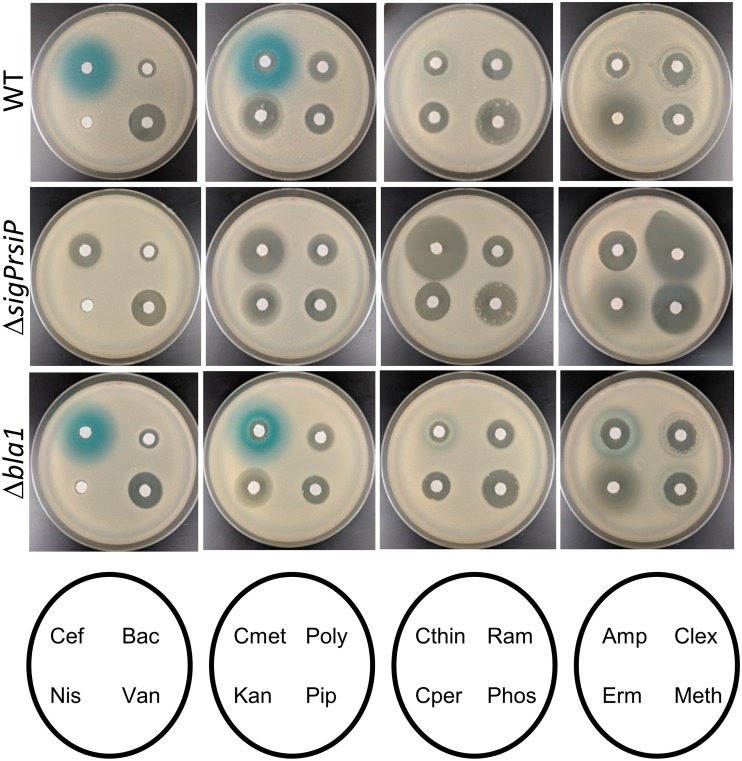
Like many ECF σ factor systems, σP is required for its own transcription (5). To monitor σP activation, we fused the σP promoter (PsigP) to the lacZ reporter gene and integrated this construct into the genome of B. thuringiensis (THE2549 thrC::PsigP-lacZ). We tested several classes of β-lactams and cell wall-targeting antibiotics for their ability to induce expression of PsigP-lacZ. We observed wide zones of PsigP-lacZ induction around cefoxitin and cefmetazole (Fig. 1). We detected fainter zones of induction in the areas around cephalothin and cephalexin (Fig. 1). Very faint zones of induction were present in the cells around ampicillin and methicillin (Fig. 1). Interestingly, we did not observe this induction surrounding the β-lactams cefoperazone and piperacillin or antibiotics that target other steps in cell wall biosynthesis, including ramoplanin, phosphomycin, nisin, bacitracin, and vancomycin (Fig. 1). We also tested compounds that do not target peptidoglycan biosynthesis, including kanamycin, polymyxin B, and erythromycin (Erm), and saw no induction of PsigP-lacZ (Fig. 1).
FIG 1.
Expression of sigP is specifically induced by β-lactams. All the strains contained PsigP-lacZ in either a wild-type (THE2549), a ΔsigP-rsiP (EBT232), or a Δbla1 (EBT215) background. Mid-log cells were washed and diluted 1:100 in molten LB agar containing X-Gal (100 μg/ml) and poured into empty 100-mm petri dishes. Filter disks containing cefoxitin (Cef) (1 μl of 5-mg/ml cefoxitin), bacitracin (Bac) (1 μl of 50-mg/ml bacitracin), nisin (Nis) (3 μl of 100-mg/ml nisin), vancomycin (Van) (1 μl of 10-mg/ml vancomycin), cefmetazole (Cmet) (1 μl of 5-mg/ml cefmetazole), polymyxin B (Poly) (1 μl of 50-mg/ml polymyxin B), kanamycin (Kan) (1 μl of 10-mg/ml kanamycin), piperacillin (Pip) (1 μl of 5-mg/ml piperacillin), cephalothin (Cthin) (1 μl of 50-mg/ml cephalothin), ramoplanin (Ram) (1 μl of 25-mg/ml ramoplanin), cefoperazone (Cper) (1 μl of 50 mg/ml cefoperazone), phosphomycin (Phos) (1 μl of 100-mg/ml phosphomycin), Amp (2 μl of 200-mg/ml ampicillin), cephalexin (Clex) (1 μl of 50-mg/ml cephalexin), Erm (1 μl of 5-mg/ml erythromycin), and methicillin (Meth) (2 μl of 100-mg/ml methicillin) were then placed on the top agar and incubated for 16 h at 30°C.
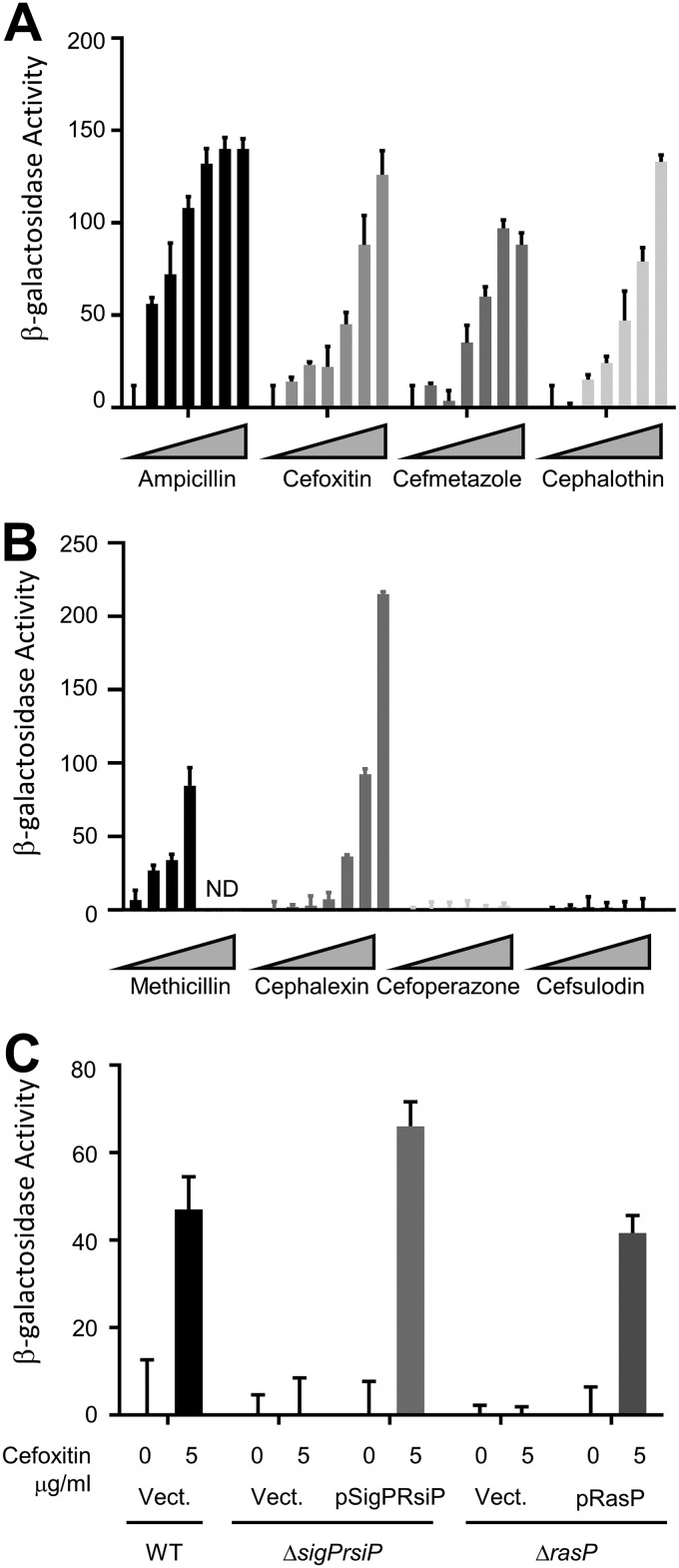
To quantify the levels of β-lactam induction, we tested eight β-lactams for their ability to activate the PsigP-lacZ fusions using a β-galactosidase assay. Mid-log cells were incubated in the presence of various concentrations of ampicillin, cefoxitin, cefmetazole, cephalothin, methicillin, cephalexin, cefoperazone, and cefsulodin for 1 h at 37°C. We observed dose-dependent induction with a subset of these β-lactams (Fig. 2A and B). Interestingly, ampicillin, methicillin, and cephalexin showed low levels of PsigP-lacZ induction when spotted onto a lawn of cells (Fig. 1) but strongly induced PsigP-lacZ in liquid assays (Fig. 2A and B), a point we will return to later. In contrast, neither cefoperazone nor cefsulodin was able to induce on the plates or in liquid (Fig. 1 and 2B). This confirms our observation that a subset of β-lactams induces σP activation.
FIG 2.
Expression of PsigP-lacZ is dose dependent and dependent upon σP and RasP. (A) B. thuringiensis with transcriptional fusion PsigP-lacZ (THE2549) was grown overnight at 30°C, subcultured in LB, and grown to an OD600 of ∼0.8 before being incubated with various concentrations of β-lactams (0, 0.0625, 0.125, 0.25 0.5, 1, and 2 μg/ml) for 1 h. Cells were collected and resuspended in Z buffer. (B) B. thuringiensis with transcriptional fusion PsigP-lacZ (THE2549) was grown overnight at 30°C, subcultured in LB, and grown to an OD600 of ∼0.8 before being incubated with various concentrations of β-lactams (0, 0.0625, 0.125, 0.25 0.5, 1, and 2 μg/ml) for 1 h. Cells were collected and resuspended in Z buffer. (C) All strains contain PsigP-lacZ and the genotype and plasmid noted: wild type/Vect. (EBT169), sigP/Vect. (EBT251), ΔsigP-rsiP/pSigPRsiP (EBT238), ΔrasP/Vect. (EBT175), and rasP/pRasP (EBT176). Strains were grown to mid-log phase and then treated with 5 μg/ml cefoxitin or untreated (0) and incubated for 1 h. β-Galactosidase activity was calculated as described in Materials and Methods. These experiments were done in triplicate, and standard deviations are represented by error bars.
We found that deletion of the sigP-rsiP genes blocked expression of PsigP-lacZ in the presence of β-lactams (Fig. 1 and 2C), demonstrating that σP is required for induction of PsigP-lacZ in response to β-lactams. When we introduced a low-copy-number plasmid containing PsigP-sigP+-rsiP+ into the ΔsigP-rsiP mutant (ΔsigP-rsiP/pSigPRsiP), we restored the induction of PsigP-lacZ in response to cefoxitin (Fig. 2C). Taken together, these data suggest that a subset of β-lactam antibiotics activates σP.
σP and Bla1 are involved in resistance to some β-lactams.
To determine the impact of σP on resistance to β-lactams, we measured the MICs of several β-lactams for wild-type and ΔsigP-rsiP mutant strains. We found that the wild type was greater than 100-fold more resistant to ampicillin, methicillin, and cephalothin than was the ΔsigP-rsiP mutant (Table 1). The wild type was 16- to 50-fold more resistant to cefmetazole, cefoxitin, and cephalexin than the mutant (Table 1). There was little or no difference in resistance to piperacillin, cefoperazone, and cefsulodin, which also failed to activate σP (Table 1 and Fig. 1). We also demonstrate that complementing the ΔsigP-rsiP mutant with a plasmid carrying PsigP-sigP+-rsiP+ restored resistance to ampicillin and cefoxitin (Table 2). For reasons that remain unclear, strains containing plasmids, including empty vector, have slight increases in β-lactam resistance. However, this does not impact the observation that the presence of PsigP-sigP+-rsiP+ restored resistance to ampicillin and cefoxitin.
TABLE 1.
ΔsigP-rsiP mutant is more sensitive to β-lactams than wild type
| Drug | MIC (μg/ml) for strain (mean ± SD): |
Fold difference | |
|---|---|---|---|
| WT | ΔsigP-rsiP mutant | ||
| Ampicillin | 6,000 ± 0 | 1.67 ± 0.5 | 3,592 |
| Cefoxitin | 200 ± 0 | 20 ± 0 | 10 |
| Methicillin | 666 ± 115 | 1 ± 0 | 666 |
| Piperacillin | 5 ± 0 | 1.25 ± 0 | 4 |
| Cephalothin | 88 ± 25 | 0.25 ± 0 | 350 |
| Cephalexin | 200 ± 0 | 4 ± 0 | 50 |
| Cefmetazole | 44 ± 13 | 2.8 ± 1.1 | 16 |
| Cefoperazone | 5 ± 2 | 4 ± 0 | 1.25 |
| Cefsulodin | 400 ± 0 | 400 ± 0 | 1 |
TABLE 2.
RasP is required for resistance to β-lactamsa
| Genotype | Vector | MIC (μg/ml) of drug (mean ± SD): |
||
|---|---|---|---|---|
| Ampicillin | Cefoxitin | Methicillin | ||
| WT | Empty | 8,000 ± 0 | 200 ± 0 | 666.7 ± 115 |
| ΔsigP-rsiP | Empty | 2 ± 0 | 20 ± 0 | 1 ± 0 |
| ΔsigP-rsiP | pSigP | 6,666 ± 3,011 | 100 ± 0 | ND |
| ΔrasP | Empty | 6.7 ± 2.1 | 20 ± 0 | ND |
| ΔrasP | pRasP | 6,333 ± 1,966 | 133 ± 57.7 | ND |
| Δbla1 | Empty | 400 ± 0 | 200 ± 0 | 125 ± 50 |
Abbreviations: WT, wild type; ND, not determined.
Since σP was shown to control expression of hd73_3490 (referred to here as bla1), which encodes a β-lactamase, we sought to determine if this gene played a role in resistance to β-lactams. We made a deletion of bla1 and determined the MIC of ampicillin and cefoxitin for this strain. The bla1 mutant was 8- to 16-fold more sensitive to ampicillin and ∼5-fold more sensitive to methicillin but no more sensitive to cefoxitin than the wild type (Table 2). This contrasts with the sigP mutant, which is greater than 1,000-fold more sensitive to ampicillin, 600-fold more sensitive to methicillin, and ∼25-fold more sensitive to cefoxitin than the wild type (Table 2). This suggests that Bla1 plays a more important role in resistance to ampicillin and methicillin than to cefoxitin. Furthermore, our data suggest that while Bla1 contributes to β-lactam resistance, additional σP-regulated genes must also contribute to β-lactam resistance.
When we tested various β-lactams for induction of PsigP-lacZ on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates, we did not consistently observe a strong zone of induction surrounding ampicillin and methicillin (Fig. 1). We hypothesized that this weak induction zone was due to the wild type efficiently producing β-lactamases which degraded the inducer (ampicillin and methicillin). Thus, we were unable to observe the increased production of β-galactosidase. To test this hypothesis, we determined the effect of a Δbla1 mutant on σP activation. We found that in the Δbla1 mutant, ampicillin and methicillin produced more distinct zones of induction (Fig. 1). However, all other induction zones of the Δbla1 mutant were similar to the wild type. Thus, in the absence of Bla1, which degrades ampicillin and methicillin, we detected greater induction of PsigP-lacZ expression. Taken together, these observations suggest that the weak ampicillin induction of PsigP-lacZ on plates is in part due to the efficient degradation of the inducer by β-lactamases.
RsiP is degraded in response to cefoxitin in a dose-dependent manner.
The anti-σ factors of other ECF01 family members are degraded, which leads to the activation of their cognate σ factors (7, 14, 15). We sought to determine if β-lactams activate σP by inducing degradation of RsiP. To investigate this, we constructed a strain with an anhydrotetracycline (ATc)-inducible copy of green fluorescent protein (GFP) fused to the N terminus of RsiP (GFP-RsiP). The inducible promoter allows us to uncouple expression of RsiP from induction of σP. The GFP-RsiP fusion allows us to follow the fate of the cytoplasmic portion of RsiP. Expression of GFP-RsiP complements an rsiP null mutation (see Fig. S1 in the supplemental material) and localizes to the membrane (Fig. S2). We then induced the synthesis of GFP-RsiP in exponential-phase cells and monitored its processing before and after treatment with cefoxitin. We chose to utilize cefoxitin for these experiments because cefoxitin induces σP activation over a wide concentration range and the ΔsigP-rsiP mutant strain grows at most of these concentrations (Fig. 2A and Table 1). Cell pellets were then lysed by sonication, and Western blot analyses were performed using anti-RsiP antisera against the extracellular portion of RsiP or anti-GFP antisera, which detect GFP fused to the intracellular portion of RsiP.
GFP-RsiP is functional. B. thuringiensis rsiP1–80 (THE2628) containing tetracycline-inducible gfp-rsiP (EBT587) or empty vector (EBT561) was plated on LB–X-Gal without or with ATc (50 ng/ml). Download FIG S1, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GFP-RsiP localizes to the membrane. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360), tetracycline-inducible gfp-rsiP1–72 (EBT533), or empty vector (EBT169) was subcultured 1:50 with ATc (100 ng/ml) and grown at 30°C to late log phase. Two microliters was spotted on a 1% agarose pad for immobilization and imaged for GFP localization. Phase-contrast and fluorescence micrographs were recorded on an Olympus BX60 microscope with a 100 UPlanApo objective (numerical aperture, 1.35). For the GFP micrographs, a filter set from Chroma Technology Corp (catalog no. 41017) was used. The GFP filter consists of a 450- to 490-nm excitation filter, a 495-nm dichroic mirror (long pass), and a 500- to 550-nm emission filter. Micrographs were captured with a Hamamatsu Orca Flash 4.0 V2 complementary metal oxide semiconductor (CMOS) camera. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
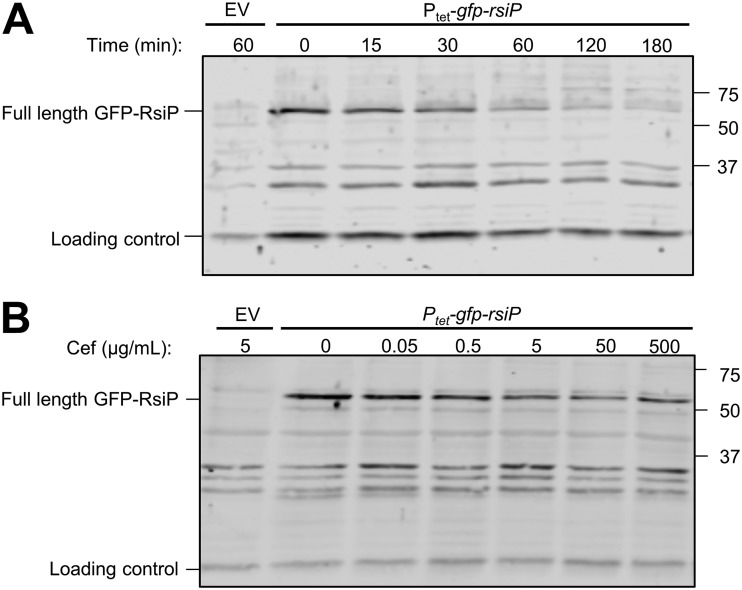
When cells producing GFP-RsiP were grown in the absence of cefoxitin, we detected full-length GFP-RsiP at the expected size of ∼60 kDa using anti-RsiP antisera. This band was absent in the empty-vector control (Fig. 3A). When cells were incubated with cefoxitin (5 μg/ml) for various times, we found that the level of full-length GFP-RsiP decreased over time (Fig. 3A and Fig. S3A). We observed loss of GFP-RsiP by 30 min to 1 h after exposure to cefoxitin (Fig. 3A and Fig. S3A). This suggests that GFP-RsiP is likely degraded in the presence of cefoxitin.
FIG 3.
RsiP levels decrease in the presence of cefoxitin. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360) or empty vector (EV; EBT169) was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cells were incubated with 5 μg/ml of cefoxitin for various times (0, 15, 30, 60, 120, or 180 min) (A) or increasing concentrations of cefoxitin (0, 0.05, 0.5, 5, 50, or 500 μg/ml) for 1 h (B). The immunoblot was probed with antisera against RsiP (α-RsiP76–275). Streptavidin IR680LT was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). The color blot showing both anti-RsiP and streptavidin on a single gel is shown in Fig. S3. Numbers at right indicate molecular masses in kilodaltons.
RsiP levels decrease in the presence of cefoxitin. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360) or empty vector (EBT169) was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cells were incubated with 5 μg/ml of cefoxitin for various times (0, 15, 30, 60, 120, or 180 min) (A) or increasing concentrations of cefoxitin (0, 0.05, 0.5, 5, 50, or 500 μg/ml) for 1 h (B). The immunoblot was probed with antisera against RsiP (α-RsiP76–275) followed by goat anti-rabbit IgG IR800CW (green). EV is wild type with pAH9 (EBT169), and GFP is wild type with pAH13 (UM20). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). Download FIG S3, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also tested the effect of cefoxitin concentration on GFP-RsiP levels by incubating cells with a range of cefoxitin concentrations (0 to 500 μg/ml) for 1 h. We found that increasing concentrations of cefoxitin resulted in a greater decrease of full-length GFP-RsiP (Fig. 3B and Fig. S3B). We obtained comparable results when we performed blotting assays for the N-terminal domain using anti-GFP antisera (Fig. S4). These data suggest that activation of σP occurs via loss of RsiP in a cefoxitin dose-dependent manner.
RsiP levels decrease in the presence of cefoxitin. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360), empty vector (EBT169), or GFP alone (UM20) was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cells were incubated with increasing concentrations of cefoxitin (0, 0.05, 0.5, 5, 50, and 500 μg/ml) for 1 h. The immunoblot was probed with antisera against GFP (α-GFP) followed by goat anti-rabbit IgG IR800CW (green). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). The Western blot is shown in black and white (A) or color (B). Download FIG S4, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RasP is necessary for σP activation.
Both σE and σW are activated by regulated intramembrane proteolysis of their cognate anti-σ factors. Proteolysis of these anti-σ factors requires multiple proteases, including the highly conserved site-2 proteases RseP and RasP, respectively (14, 15). We hypothesize that activation of σP requires multiple proteases, including the conserved site-2 protease RasP to degrade RsiP. To test this, we used BLAST to identify a putative membrane-embedded metalloprotease, HD73_4103, which is 76% similar and 60% identical to B. subtilis RasP and is here referred to as RasP (Fig. S5) (38–43). To determine if RasP was required for σP activation, we generated a strain containing a deletion of rasP and the PsigP-lacZ reporter. In the absence of RasP, we did not detect increased expression of PsigP-lacZ reporter in response to cefoxitin (Fig. 2C). In MIC experiments, we found that, similarly to the ΔsigP-rsiP mutant, the ΔrasP mutant was more sensitive to ampicillin and cefoxitin (Table 2). We found that both resistance to β-lactams and induction of PsigP-lacZ could be complemented when a plasmid expressing rasP+ was introduced into the ΔrasP mutant (Fig. 2C and Table 2). These data suggest that RasP is required for σP activation.
Amino acid alignment of RasP. An alignment of B. subtilis RasP and B. thuringiensis HD73_4301. The active site is marked by a red box. Download FIG S5, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RasP is required for degradation of RsiP.
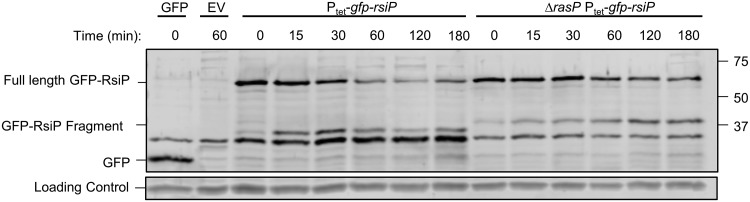
To determine if RasP is required for degradation of RsiP, we expressed the GFP-RsiP fusion in both the wild type and a ΔrasP mutant. We treated cells with 5 μg/ml cefoxitin for various lengths of time from 0 to 180 min (Fig. 4 and Fig. S6). In the wild type, we observed loss of full-length RsiP over time (Fig. 4 and Fig. S6). In contrast, we observed loss of full-length GFP-RsiP and the accumulation of a smaller ∼35-kDa band in the ΔrasP mutant (Fig. 4 and Fig. S6). This suggests that RasP is required for complete degradation of RsiP. Since a truncated product accumulates in the ΔrasP mutant, RasP is likely required for site-2 cleavage and an unidentified protease is required for cleavage at site-1.
FIG 4.
RsiP degradation is dependent upon the site-2 protease RasP. B. thuringiensis wild type (EBT360) or ΔrasP (EBT366) containing a tetracycline-inducible copy of gfp-rsiP was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cultures were incubated with cefoxitin (5 μg/ml) for the time indicated at 37°C. The immunoblot was probed with anti-GFP antisera. EV is wild type with pAH9 (EBT169), and GFP is wild type with pAH13 (UM20). Streptavidin IR680LT was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). The color blot showing both anti-GFP and streptavidin on a single gel is shown in Fig. S6. Numbers at right are molecular masses in kilodaltons.
RsiP degradation is dependent upon the site-2 protease RasP. B. thuringiensis containing a tetracycline-inducible copy of gfp-rsiP, wild type (EBT360) or ΔrasP (EBT366), was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cultures were incubated for 1 h without (−) or with (+) cefoxitin treatment (5 μg/ml) at 37°C. The immunoblot was probed with antisera against GFP (anti-GFP) followed by goat anti-rabbit IgG IR800CW (green). EV is wild type with pAH9 (EBT169), and GFP is wild type with pAH13 (UM20). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). Download FIG S6, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutations in rsiP result in constitutive sigP expression.
To further characterize the σP signal transduction system, we isolated mutants which resulted in constitutive expression of PsigP-lacZ. We selected for mutants with increased resistance to cefoxitin by plating cultures of the wild-type PsigP-lacZ strain (THE2549) on LB-cefoxitin (200 μg/ml) agar. At this concentration of cefoxitin, wild-type B. thuringiensis fails to grow. These strains were tested for PsigP-lacZ expression in the absence of cefoxitin by streaking on LB–X-Gal. We isolated 8 independent mutants with increased resistance to cefoxitin that have constitutive PsigP-lacZ expression. We hypothesized that these strains harbored mutations in rsiP. We PCR amplified and sequenced the sigP and rsiP genes from the constitutive mutants. The 8 constitutive mutants contained mutations in different regions of the rsiP gene that resulted in C-terminal truncations of RsiP (Fig. S7). We selected four rsiP mutants for further study. We found that each mutant strain showed increased PsigP-lacZ expression even in the absence of β-lactams (Fig. 5). When a wild-type copy of rsiP (pSigPRsiP) was introduced to each of these mutants, PsigP-lacZ expression was no longer constitutive but was induced in the presence of cefoxitin (Fig. S8). This indicates that the rsiP mutations were responsible for the increased PsigP-lacZ expression.
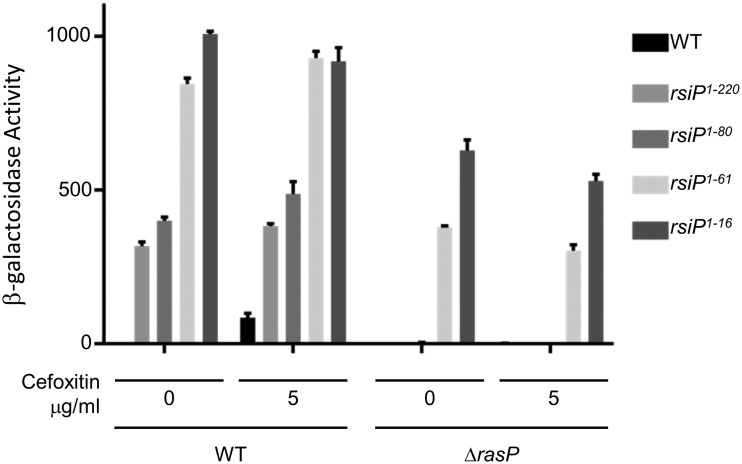
FIG 5.
Truncations of RsiP lead to constitutive σP activation. To determine if RasP was required for ampicillin-inducible PsigP-lacZ expression, we assayed β-galactosidase activity of B. thuringiensis with transcriptional fusion PsigP-lacZ and different rsiP truncation mutants (WT, THE2549; RsiP1–220, THE2602; RsiP1–80, THE2628; RsiP1–61, THE2637; RsiP1–16, THE2642) and a ΔrasP deletion (WT, EBT140; RsiP1–220, EBT116; RsiP1–80, EBT148; RsiP1–61, EBT133; RsiP1–16, THE2605). Cells were grown overnight at 30°C, subcultured in LB, and grown to an OD600 of ∼0.8 before being incubated with cefoxitin (5 μg/ml) for 1 h. The experiment was performed in triplicate, and standard deviations are represented by error bars.
RsiP mutants result in constitutive σP activity. Alignment of RsiP mutants that result in nonsense, frameshift, or point mutations. Amino acid residues in red are indicative of the change in amino acid sequence due to mutations. The mutation rsiP1–232 was isolated 3 independent times while rsiP1–61 and rsiP1–15 were isolated twice each. The red box indicates the predicted transmembrane domain. Download FIG S7, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of RsiP mutants. B. thuringiensis containing either empty vector (wild type, EBT169; rsiP1–220, EBT564; rsiP1–80, EBT565; rsiP1–61, EBT566; rsiP1–16, EBT567) or PsigP-sigP-rsiP (wild type, EBT168; rsiP1–220, EBT560; rsiP1–80, EBT651; rsiP1–61, EBT562; rsiP1–16, EBT563) was grown overnight and spotted onto LB–X-Gal (100 μg/ml) plates lacking cefoxitin (A) or containing cefoxitin (5 μg/ml) (B). Download FIG S8, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the σV and σW systems, RasP cleaves the anti-σ factors RsiW and RsiV within the transmembrane domain to activate the cognate σ factors (15, 35). The RsiP transmembrane is predicted to be residues 54 to 71 based on TMHMM (44). Two of the four RsiP truncations produce proteins with the transmembrane domain intact, while the remaining RsiP truncations lack the transmembrane domain. Since RasP is known to cleave proteins within the transmembrane domain, we hypothesized that those truncations which still contain a transmembrane domain would require RasP in order to activate σP. To test this, we introduced the ΔrasP mutation into each of the rsiP mutants. In the absence of RasP, strains containing truncations which have a transmembrane domain (RsiP1–220 and RsiP1–80) (Fig. 4 and Fig. S7) no longer constitutively activate σP (Fig. 5). However, the strains with the rsiP truncation lacking the transmembrane domain (RsiP1–16 and RsiP1–61) constitutively activate σP even in the absence of RasP (RsiP1–16 and RsiP1–61) (Fig. 4 and Fig. S5). Thus, RasP is required for σP activation when the transmembrane domain of RsiP is intact, consistent with the role of RasP as a site-2 protease.
RasP cleaves within the transmembrane domain of RsiP and is not the regulated step in σP activation.
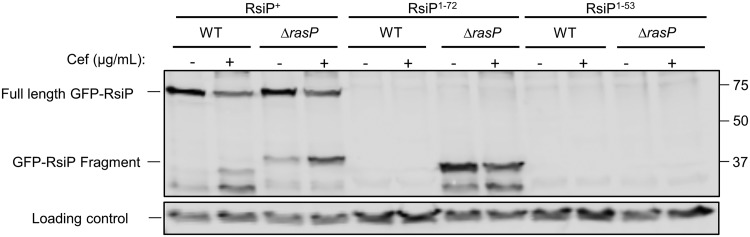
In the case of σW and σV, the rate-limiting step in σ factor activation is site-1 cleavage (15, 35). Since the identity of the site-1 protease is not currently known, we sought to determine if RasP cleavage of RsiP is a rate-limiting step in σP activation. To test this, we constructed truncations of GFP-RsiP that lack the extracellular portion of RsiP. One truncation includes the transmembrane domain (gfp-rsiP1–72), and one truncation lacks the transmembrane domain (gfp-rsiP1–53). We expressed the truncated GFP-RsiP proteins in wild-type and ΔrasP backgrounds and exposed these strains to cefoxitin (5 μg/ml). In wild-type strains, we found that both GFP-RsiP1–72 and GFP-RsiP1–53 were degraded (Fig. 6 and Fig. S9). However, in the ΔrasP mutant GFP-RsiP1–72 accumulated, while GFP-RsiP1–53 was degraded (Fig. 6 and Fig. S9). These data indicate that GFP-RsiP1–72 requires RasP for degradation while GFP-RsiP1–53 does not. One possible interpretation is that GFP-RsiP1–72 is not produced or localized properly to the membrane. Thus, we confirmed that GFP-RsiP1–72 localizes to the membrane by fluorescence microscopy (Fig. S2). This suggests that the RasP cleavage site of RsiP occurs within the transmembrane domain between amino acids 53 and 72. The presence or absence of cefoxitin had no effect on the degradation (Fig. 6 and Fig. S9). Since GFP-RsiP1–72 is constitutively degraded, we conclude that GFP-RsiP1–72 mimics the site-1 cleavage product and that RasP activity is not induced by cefoxitin. This suggests that RasP cleavage of RsiP is not the regulated step in σP activation and that site-1 cleavage is the step that is controlled by the presence of β-lactams.
FIG 6.
Truncation of RsiP results in constitutive degradation in a RasP-dependent manner. B. thuringiensis containing a tetracycline-inducible copy of gfp-rsiP, gfp-rsiP1–72 (rsiP without the extracellular domain), or gfp-rsiP1–53 (rsiP without the transmembrane and extracellular domains) was constructed in either the wild type (rasP+) or a ΔrasP mutant strain (GFP-RsiP wild type [rasP+], EBT360; GFP-RsiP ΔrasP, EBT366; GFP-RsiP1–53 wild type [rasP+], EBT518; GFP-RsiP1–53 ΔrasP, EBT510; GFP-RsiP1–72 wild type [rasP+], EBT516; GFP-RsiP1–72 ΔrasP, EBT533). Strains were subcultured 1:50 into LB supplemented with ATc (100 ng/ml), grown to mid-log phase, and then incubated for 2 h without (−) or with (+) cefoxitin treatment (5 μg/ml) at 37°C. The immunoblot was probed with anti-GFP antisera. Streptavidin IR680LT was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). The color blot showing both anti-GFP and streptavidin on a single gel is shown in Fig. S9. Numbers at right are molecular masses in kilodaltons.
Truncation of RsiP results in constitutive degradation in a RasP-dependent manner. B. thuringiensis containing a tetracycline-inducible copy of gfp-rsiP, gfp-rsiP1–72 (rsiP without the extracellular domain) or gfp-rsiP1–53 (rsiP without the transmembrane and extracellular domains), was constructed in either wild type or a ΔrasP mutant strain (GFP-RsiP wild type [rasP+], EBT360; GFP-RsiP ΔrasP, EBT366; GFP-RsiP1–53 wild type [rasP+], EBT518; GFP-RsiP1–53 ΔrasP, EBT510; GFP-RsiP1–72 wild type [rasP+], EBT516; GFP-RsiP1–72 ΔrasP, EBT533). Strains were subcultured 1:50 into LB supplemented with ATc (100 ng/ml) and at mid-log phase were incubated for 2 h without (−) or with (+) cefoxitin treatment (5 μg/ml) at 37°C. The immunoblot was probed with antisera against GFP (anti-GFP) followed by goat anti-rabbit IgG IR800CW (green). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). Download FIG S9, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Many ECF σ factors are induced in response to extracytoplasmic stressors and initiate transcription of a subset of genes to modulate the cell’s response to these stresses. ECF σ factors can respond to signals such as misfolded periplasmic protein, antimicrobial peptides, or lysozyme. The ECF σ factors encoded in highly related organisms can vary widely. For example, B. subtilis encodes 7 ECF σ factors, while B. thuringiensis encodes 15 predicted ECF σ factors. The only ECF σ factor that these organisms share is σM (45). Thus, there is a variability in how bacteria utilize ECF σ factors to respond to stress. Ross et al. demonstrated that the novel ECF σ factor σP is induced in the presence of ampicillin and initiates transcription of β-lactamases (5). Here, we demonstrated that σP responds specifically to a subset of β-lactams, while other β-lactams and cell wall-targeting antibiotics fail to induce σP activation. We also showed that σP confers various degrees of resistance to these β-lactam antibiotics. We found that σP was not required for resistance to other cell wall antibiotics, including vancomycin, nisin, and bacitracin, suggesting specificity in resistance to β-lactams and not a general cell envelope stress response.
For ECF σ factors to be activated, their cognate anti-σ factors must be inactivated. This can be accomplished via various mechanisms, including a conformational change of the anti-σ factor; partner switching, where an anti-anti-σ factor frees the σ factor from the anti-σ factor; or proteolytic destruction of the anti-σ factor (9, 11). The anti-σ factors RseA in E. coli and RsiW and RsiV in B. subtilis are degraded sequentially by regulated intramembrane proteolysis. Each of these anti-σ factors requires a different family of proteases to cleave the anti-σ factor at site-1 (14, 22, 30, 46, 47), while site-2 cleavage is carried out by the conserved site-2 protease (14, 15, 35). We hypothesize that σP is activated in a similar manner. Our data indicate that σP is released from RsiP by proteolytic degradation when β-lactams are present. We found that RasP is required for activation of σP. We also observe that an RsiP degradation product approximately the size of our predicted RasP substrate accumulates in a ΔrasP mutant. This indicates that RasP is required for degradation of RsiP. Our data also suggest, similarly to other anti-σ factors, that site-2 cleavage of RsiP is not the rate-limiting step, since the C-terminal RsiP truncations are constitutively degraded and lead to constitutive σP activation in the absence of β-lactams. Thus, we hypothesize that RasP is required for site-2 cleavage of RsiP and that an as-yet-unidentified protease is required to initiate degradation of RsiP by cleaving RsiP at site-1. We hypothesize that, like other ECF σ factors activated by regulated intramembrane proteolysis, site-1 cleavage of RsiP is likely the rate-limiting step in σP activation.
Our data suggest that a subset of β-lactams induce σP activation. We found that, in addition to ampicillin, σP is activated by cefoxitin, cefmetazole, cephalothin, cephalexin, and methicillin but not by piperacillin, cefoperazone, cefsulodin, or antibiotics that target other steps in peptidoglycan biosynthesis. This raises the question of what the signal is for σP activation. The β-lactams could be sensed directly or indirectly. For example, RsiV directly senses lysozyme and degradation of RsiV is rapid (31). In contrast, activation of σE is indirect and due to buildup of products that occur when the outer membrane is damaged (31, 48). Our data suggest that RsiP degradation is a relatively slow process. One possible interpretation of this is that β-lactam-induced peptidoglycan (PG) damage must accumulate to induce RsiP degradation. We hypothesize that the β-lactams that we tested have different affinities for penicillin-binding proteins (PBPs) and that this affinity may explain why some β-lactams induce σP while others do not. In other organisms, including Streptococcus pneumoniae, B. subtilis, and E. coli, β-lactams can differentially target PBPs (49–51). This raises the possibility that activation of σP could be the result of inhibition of specific PBPs. Unfortunately, at this time we do not know which PBPs are targeted by the different β-lactams in B. thuringiensis. Thus, the precise mechanism and signal responsible for σP activation remain to be clearly defined.
MATERIALS AND METHODS
Media and growth conditions.
All B. thuringiensis strains are isogenic derivatives of AW43, a derivative of Bacillus thuringiensis subsp. kurstaki strain HD73 (52). All strains and genotypes can be found in Table 3. All B. thuringiensis strains were grown in or on LB medium at 30°C unless otherwise specified. Cultures of B. thuringiensis were grown with agitation in a roller drum. Strains containing episomal plasmids were grown in LB containing chloramphenicol (Cam; 10 μg/ml) or erythromycin (Erm; 10 μg/ml). E. coli strains were grown at 37°C using LB-ampicillin (Amp; 100 μg/ml) or LB-Cam (10 μg/ml) medium. To screen for threonine auxotrophy, B. thuringiensis strains were patched on minimal medium plates without or with threonine (50 μg/ml) (53, 54). The β-galactosidase chromogenic indicator 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at a concentration of 100 μg/ml. Anhydrotetracycline (ATc; Sigma) was used at a concentration of 100 ng/ml.
TABLE 3.
Strains
| Species and strain | Description | Reference or source |
|---|---|---|
| B. thuringiensis | ||
| AW43 |
B. thuringiensis subsp. kurstaki HD73 cured of both pAW63 and pHT73, Nalr |
52 |
| THE2549 | AW43 thrC::PsigP-lacZ | This study |
| EBT140 | AW43 thrC::PsigP-lacZ ΔrasP | This study |
| EBT232 | AW43 thrC::PsigP-lacZ ΔsigP-rsiP | This study |
| EBT215 | AW43 thrC::PsigP-lacZ Δbla1 | This study |
| EBT360 | AW43 thrC::PsigP-lacZ/pAH9 Ptet-gfp-rsiP | This study |
| EBT366 | AW43 thrC::PsigP-lacZ ΔrasP/pAH9 Ptet-gfp-rsiP | This study |
| EBT510 | AW43 thrC::PsigP-lacZ ΔrasP/pAH9 Ptet-gfp-rsiP1–53 | This study |
| EBT516 | AW43 thrC::PsigP-lacZ/pAH9 Ptet-gfp-rsiP1–72 | This study |
| EBT518 | AW43 thrC::PsigP-lacZ/pAH9 Ptet-gfp-rsiP1–53 | This study |
| EBT533 | AW43 thrC::PsigP-lacZ ΔrasP/pAH9 Ptet-gfp-rsiP1–72 | This study |
| EBT175 | AW43 thrC::PsigP-lacZ ΔrasP/pAH9 | This study |
| EBT176 | AW43 thrC::PsigP-lacZ ΔrasP/pAH9 rasP | This study |
| EBT238 | AW43 thrC::PsigP-lacZ ΔsigP-rsiP/pAH9 PsigP-sigP-rsiP | This study |
| EBT251 | AW43 thrC::PsigP-lacZ ΔsigP-rsiP/pAH9 | This study |
| THE2642 | AW43 thrC::PsigP-lacZ rsiP1–16 | This study |
| THE2637 | AW43 thrC::PsigP-lacZ rsiP1–61 | This study |
| THE2628 | AW43 thrC::PsigP-lacZ rsiP1–80 | This study |
| THE2602 | AW43 thrC::PsigP-lacZ rsiP1–220 | This study |
| THE2605 | AW43 thrC::PsigP-lacZ ΔrasP rsiP1–16 | This study |
| EBT133 | AW43 thrC::PsigP-lacZ ΔrasP rsiP1–61 | This study |
| EBT148 | AW43 thrC::PsigP-lacZ ΔrasP rsiP1–80 | This study |
| EBT116 | AW43 thrC::PsigP-lacZ ΔrasP rsiP1–220 | This study |
| EBT567 | AW43 thrC::PsigP-lacZ rsiP1–16/pAH9 PsigP-sigP-rsiP | This study |
| EBT566 | AW43 thrC::PsigP-lacZ rsiP1–61/pAH9 PsigP-sigP-rsiP | This study |
| EBT565 | AW43 thrC::PsigP-lacZ rsiP1–80/pAH9 PsigP-sigP-rsiP | This study |
| EBT564 | AW43 thrC::PsigP-lacZ rsiP1–220/pAH9 PsigP-sigP-rsiP | This study |
| EBT168 | AW43 thrC::PsigP-lacZ/pAH9 PsigP-sigP-rsiP | This study |
| EBT169 | AW43 thrC::PsigP-lacZ pAH9 | This study |
| EBT563 | AW43 thrC::PsigP-lacZ rsiP1–16/pAH9 | This study |
| EBT562 | AW43 thrC::PsigP-lacZ rsiP1–61/pAH9 | This study |
| EBT561 | AW43 thrC::PsigP-lacZ rsiP1–80/pAH9 | This study |
| EBT560 | AW43 thrC::PsigP-lacZ rsiP1–220/pAH9 | This study |
| UM20 | AW43/pAH13 | This study |
| EBT587 | AW43 thrC::PsigP-lacZ rsiP1–80/pAH9 Ptet-gfp-rsiP | This study |
| E. coli | ||
| OmniMax 2-T1R | F′ {proAB+
lacIq
lacZΔM15 Tn10(Tetr) Δ(ccdAB)} mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80(lacZ)ΔM15 Δ(lacZYA-argF)U169 endA1 recA1 supE44 thi-1 gyrA96 relA1 tonA panD |
Invitrogen |
| INV110 |
endA1 rpsL thr leu thi lacY galK galT ara tomA tsx dam dcm supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] |
Invitrogen |
Strain and plasmid construction.
All plasmids are listed in Table 4, which includes information relevant to plasmid assembly. Plasmids were constructed by isothermal assembly (55). Regions of plasmids constructed using PCR were verified by DNA sequencing. The oligonucleotide primers used in this work were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Table S1 in the supplemental material. All plasmids were propagated using OmniMax 2-T1R as the cloning host and passaged through the nonmethylating E. coli strain INV110 before being transformed into a B. thuringiensis recipient strain.
TABLE 4.
Plasmids
| Plasmid | Relevant feature(s) | Parent vector | Digestion enzymes | Insert primers | Reference |
|---|---|---|---|---|---|
| pMAD | ori-pE194ts | 56 | |||
| pAH9 | ori-pE194 PsarA-mcherry | 57 | |||
| pAH13 | Ptet-gfp | 57 | |||
| pRAN332 | Ptet-gfp | 64 | |||
| pEBT4 | ori-pE194ts, ΔblaP | pMAD | BgIII, EcoRI | 3832 and 3833, 3834 and 3835 | This study |
| pEBT5 | ori-pE194ts, ΔrasP | pMAD | BgIII, EcoRI | 3632 and 3633, 3634 and 3635 | This study |
| pEBT6 | ori-pE194ts, ΔsigP-rsiP | pMAD | BgIII, EcoRI | 3776 and 3777, 3778 and 3779 | This study |
| pEBT13 | Ptet-gfp-rsiP | pAH9 | HindIII, EcoRI | 3838 and 3839 | This study |
| pCE630 | Ptet-gfp-rsiP1–72 | pAH9 | HindIII, EcoRI | 3838 and 4258 | This study |
| pCE632 | Ptet-gfp-rsiP1–53 | pAH9 | HindIII, EcoRI | 3838 and 4259 | This study |
| pTHE960 | PsigP-sigP+-rsiP+ | pAH9 | HindIII, EcoRI | 3774 and 3775 | This study |
| pIA02 | PsarA-rasP+ | pAH9 | EcoRI, KpnI | 3744 and 3745 | This study |
| pTHE946 | pE194ts | pMAD | BamHI, StuI | This study | |
| pTHE948 | pE194ts ‘thrC thrB’ | pTHE946 | ScaI, SalI | 2917 and 2918, 2919 and 2920 | This study |
| pTHE950 | pE194ts ‘thrC lacZ thrB’ | pTHE948 | XhoI, SbfI | 2922 and 2923 | This study |
| pTHE949 | pE194ts ‘thrC PsigP-lacZ thrB’ | pTHE950 | XhoI, SalI | 2929 and 2930 | This study |
Oligonucleotides. Download Table S1, PDF file, 0.06 MB (65.5KB, pdf) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To construct deletion mutants, we cloned DNA 1 kb upstream and 1 kb downstream of the site of desired deletion using primers listed in Table S1 onto the temperature-sensitive pMAD plasmid (erythromycin resistant) between the BglII and EcoRI sites (56).
Complementation constructs were constructed in pAH9, which is an E. coli–Gram-positive bacterial shuttle vector with a pE194 origin of replication (57). Chromosomal DNA including the promoter sequence was cloned for PsigP-sigP+-rsiP+ and cloned into pAH9 digested with EcoRI and HindIII, while rasP was cloned downstream of the PsarA promoter from Staphylococcus aureus by digesting with EcoRI and KpnI. In B. thuringiensis, PsarA has moderate constitutive expression.
To generate strains containing the sigP promoter fused to the lacZ reporter integrated into the chromosome, we constructed a number of intermediate vectors. To switch the antibiotic resistance of the temperature-sensitive pMAD vector, we constructed pTHE946, which contains the E. coli origin (ColE1 ori) of replication, an Erm resistance gene (for selection in Gram-positive bacteria), an Amp resistance gene (for selection in E. coli strains), and the temperature-sensitive origin (pE194 ori) from pMAD (7.3-kb StuI and BamHI fragment) as well as the conjugation origin of transfer and the Cam resistance gene from pRPF185 (SmaI and BamHI fragment). The thrC (primers 2917 and 2918) and thrB (primers 2919 and 2920) genes were cloned into the ScaI- and SalI-digested pTHE946 plasmid (lacking Ermr and Ampr genes) to generate a vector (pTHE948) which can integrate into the thrC operon. A promoterless lacZ fragment (primers 2922 and 2923) was added between the thrC and thrB genes of pTHE948 (XhoI and SbfI) to generate pTHE950. This plasmid (XhoI and NotI digested) was used to clone the sigP promoter (primers TE2929 and 2930) to generate the PsigP-lacZ promoter fusion (pTHE949).
B. thuringiensis DNA transformation.
Plasmids were introduced into B. thuringiensis by electroporation (58, 59). Briefly, recipient cells were grown to late log phase at 37°C. For each transformation, cells (1.5 ml) were pelleted by centrifugation (9,000 × g) and washed twice in room-temperature sterile water. After careful removal of all residual water, 100 μl of sterile 40% polyethylene glycol (PEG) 6000 (Sigma) was used to gently resuspend cells. Approximately 2 to 10 μl of unmethylated DNA (>50 ng/μl) was added to cells and transferred to an 0.4-cm-gap electroporation cuvette (Bio-Rad). Cells were exposed to 2.5 kV for 4 to 6 ms. LB was immediately added, and cells were incubated at 30°C for 1 to 2 h prior to plating on selective media.
Construction of deletions or promoter-lacZ fusions in B. thuringiensis.
To generate unmarked mutants and thrC::PsigP-lacZ strains, we used plasmid vectors containing the temperature-sensitive origin of replication (pE194 ori) from the pMAD plasmid (56). At permissive temperatures (30°C), pMAD replicates episomally as a plasmid. At nonpermissive temperatures (42°C), pMAD must integrate into the chromosome via homologous recombination; otherwise, the plasmid will be lost to segregation and the strain will become sensitive to erythromycin. Plasmids were transformed into a B. thuringiensis recipient strain and selected for on LB-Erm agar at 30°C. To select for the integration of the deletion plasmid into the recipient strain genome, plasmid-containing bacteria were grown at 42°C on LB-Erm plates. The plasmid-integrated strain was then struck on LB agar at 30°C twice. Individual colonies were patched on LB and LB-Erm agar to identify the Erm-sensitive bacteria which had lost the deletion plasmid by segregation. To verify each deletion, genomic DNA was isolated from each strain candidate and PCR was used to verify the deletion. Integration of the PsigP-lacZ fusion into the thrC operon results in threonine auxotrophy and can be identified by lack of growth on minimal medium plates without threonine.
Zones of inhibition and zones of induction.
To determine the zones of inhibition and induction by various antibiotics, we first washed mid-logarithmically grown cells in fresh LB. Washed cells were diluted 1:100 in molten LB agar containing X-Gal (100 μg/ml) and poured into empty 100-mm petri dishes. Sterile cellulose disks (8 mm) were saturated with different antibiotics and allowed to dry for longer than 10 min. After each antibiotic disk was placed on the solidified agar, plates were incubated at 30°C overnight before observation.
β-Galactosidase assays.
To quantify expression from the sigP promoter, we measured the β-galactosidase activity of cells containing a PsigP-lacZ promoter fusion. Overnight cultures were diluted 1:50 in fresh LB medium and incubated for 3 h at 30°C. One milliliter of each subculture was pelleted (9,000 × g), washed (in LB broth), and resuspended in 1 ml LB broth lacking or including specified antibiotics. After 1 h of incubation at 37°C, 1 ml of each sample was pelleted and resuspended in 200 μl of Z buffer. Cells were permeabilized by mixing with 16 μl chloroform and 16 μl 2% Sarkosyl (26, 60). Permeabilized cells (100 μl) were mixed with 10 mg/ml ortho-nitrophenyl-β-galactoside (ONPG; Research Products International; 50 μl), and optical density at 405 nm (OD405) was measured over time using an Infinite M200 Pro plate reader (Tecan). β-Galactosidase activity units (μmol of ONP formed min−1) × 103/(OD600 × ml of cell suspension) were calculated as previously described (61). Experiments were performed in triplicate with the mean and standard deviation being shown.
MIC assay.
To determine the MICs of various antibiotics, we diluted overnight cultures of bacteria (washed in LB) 1:1,000 in medium containing 2-fold dilutions of each antibiotic. All MIC experiments were performed in round-bottom 96-well plates. Each experiment was performed in triplicate, and the plates were allowed to incubate for 24 h at 37°C before observation of growth or no growth.
Immunoblot analysis.
Samples were electrophoresed on a 15% SDS-polyacrylamide gel, and proteins were then blotted onto a nitrocellulose membrane (GE Healthcare, Amersham). Nitrocellulose was blocked with 5% bovine serum albumin (BSA), and proteins were detected with either 1:10,000 anti-GFP or 1:5,000 anti-RsiP76–275 antiserum. Streptavidin IR680LT (1:10,000) was used to detect two biotin-containing proteins, PycA (HD73_4231) and AccB (HD73_4487), which served as loading controls (62, 63). To detect primary antibodies, the blots were incubated with 1:10,000 goat anti-rabbit IR800CW (Li-Cor) and imaged on an Odyssey CLx scanner (Li-Cor) or an Azure Sapphire imager (Azure Biosystems). All immunoblot assays were performed a minimum of three times with a representative example being shown.
ACKNOWLEDGMENTS
This work was supported by the Department of Microbiology and Immunology at the University of Iowa and by NIH grant R21AI146769 to C.D.E.
We thank Theresa Koehler for strains and advice. We also thank Leyla Slamti for protocols and members of the Ellermeier and Weiss labs for helpful comments.
REFERENCES
- 1.Waxman DJ, Strominger JL. 1983. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem 52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 2.Tomasz A. 1979. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol 33:113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Succi J, Tenover FC, Koehler TM. 2003. β-Lactamase genes of the penicillin-susceptible Bacillus anthracis Sterne strain. J Bacteriol 185:823–830. doi: 10.1128/JB.185.3.823-830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Tenover FC, Koehler TM. 2004. β-Lactamase gene expression in a penicillin-resistant Bacillus anthracis strain. Antimicrob Agents Chemother 48:4873–4877. doi: 10.1128/AAC.48.12.4873-4877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross CL, Thomason KS, Koehler TM. 2009. An extracytoplasmic function sigma factor controls beta-lactamase gene expression in Bacillus anthracis and other Bacillus cereus group species. J Bacteriol 191:6683–6693. doi: 10.1128/JB.00691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gargis AS, McLaughlin HP, Conley AB, Lascols C, Michel PA, Gee JE, Marston CK, Kolton CB, Rodriguez-R LM, Hoffmaster AR, Weigel LM, Sue D. 2018. Analysis of whole-genome sequences for the prediction of penicillin resistance and β-lactamase activity in Bacillus anthracis. mSystems 3:e00154-18. doi: 10.1128/mSystems.00154-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 8.Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110. doi: 10.1016/S0065-2911(02)46002-X. [DOI] [PubMed] [Google Scholar]
- 9.Helmann JD. 2016. Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol 30:122–132. doi: 10.1016/j.mib.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helmann JD. 1999. Anti-sigma factors. Curr Opin Microbiol 2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 11.Ho TD, Ellermeier CD. 2012. Extra cytoplasmic function σ factor activation. Curr Opin Microbiol 15:182–188. doi: 10.1016/j.mib.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sineva E, Savkina M, Ades SE. 2017. Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr Opin Microbiol 36:128–137. doi: 10.1016/j.mib.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshimura M, Asai K, Sadaie Y, Yoshikawa H. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591–599. doi: 10.1099/mic.0.26712-0. [DOI] [PubMed] [Google Scholar]
- 14.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schöbel S, Zellmeier S, Schumann W, Wiegert T. 2004. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol 52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 16.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. 2013. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ades SE, Connolly LE, Alba BM, Gross CA. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-σ factor. Genes Dev 13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 19.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. 2004. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 20.Kanehara K, Ito K, Akiyama Y. 2003. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J 22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama Y, Kanehara K, Ito K. 2004. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J 23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellermeier CD, Losick R. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev 20:1911–1922. doi: 10.1101/gad.1440606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butcher BG, Helmann JD. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol Microbiol 60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 24.Pietiäinen M, Gardemeister M, Mecklin M, Leskelä S, Sarvas M, Kontinen VP. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577–1592. doi: 10.1099/mic.0.27761-0. [DOI] [PubMed] [Google Scholar]
- 25.Benachour A, Muller C, Dabrowski-Coton M, Le Breton Y, Giard J-C, Rincé A, Auffray Y, Hartke A. 2005. The Enterococcus faecalis SigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments. J Bacteriol 187:1022–1035. doi: 10.1128/JB.187.3.1022-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho TD, Hastie JL, Intile PJ, Ellermeier CD. 2011. The Bacillus subtilis extracytoplasmic function sigma factor sigma(V) is induced by lysozyme and provides resistance to lysozyme. J Bacteriol 193:6215–6222. doi: 10.1128/JB.05467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guariglia-Oropeza V, Helmann JD. 2011. Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J Bacteriol 193:6223–6232. doi: 10.1128/JB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho TD, Williams KB, Chen Y, Helm RF, Popham DL, Ellermeier CD. 2014. Clostridium difficile extracytoplasmic function σ factor σV regulates lysozyme resistance and is necessary for pathogenesis in the hamster model of infection. Infect Immun 82:2345–2355. doi: 10.1128/IAI.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho TD, Ellermeier CD. 2019. Activation of the extracytoplasmic function σ factor σV by lysozyme. Mol Microbiol doi: 10.1111/mmi.14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastie JL, Williams KB, Sepúlveda C, Houtman JC, Forest KT, Ellermeier CD. 2014. Evidence of a bacterial receptor for lysozyme: binding of lysozyme to the anti-σ factor RsiV controls activation of the ECF σ factor σV. PLoS Genet 10:e1004643. doi: 10.1371/journal.pgen.1004643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastie JL, Williams KB, Bohr LL, Houtman JC, Gakhar L, Ellermeier CD. 2016. The anti-sigma factor RsiV is a bacterial receptor for lysozyme: co-crystal structure determination and demonstration that binding of lysozyme to RsiV is required for σV activation. PLoS Genet 12:e1006287. doi: 10.1371/journal.pgen.1006287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. 2002. Signal peptidases. Chem Rev 102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 33.Tuteja R. 2005. Type I signal peptidase: an overview. Arch Biochem Biophys 441:107–111. doi: 10.1016/j.abb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 34.van Roosmalen ML, Geukens N, Jongbloed JDH, Tjalsma H, Dubois J-Y, Bron S, van Dijl JM, Anné J. 2004. Type I signal peptidases of Gram-positive bacteria. Biochim Biophys Acta 1694:279–297. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Hastie JL, Williams KB, Ellermeier CD. 2013. The activity of σV, an extracytoplasmic function σ factor of Bacillus subtilis, is controlled by a regulated proteolysis of the anti-σ factor RsiV. J Bacteriol 195:3135–3144. doi: 10.1128/JB.00292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingston AW, Liao X, Helmann JD. 2013. Contributions of the σ(W), σ(M) and σ(X) regulons to the lantibiotic resistome of Bacillus subtilis. Mol Microbiol 90:502–518. doi: 10.1111/mmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jervis AJ, Thackray PD, Houston CW, Horsburgh MJ, Moir A. 2007. SigM-responsive genes of Bacillus subtilis and their promoters. J Bacteriol 189:4534–4538. doi: 10.1128/JB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider JS, Glickman MS. 2013. Function of site-2 proteases in bacteria and bacterial pathogens. Biochim Biophys Acta 1828:2808–2814. doi: 10.1016/j.bbamem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinch LN, Ginalski K, Grishin NV. 2006. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci 15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroos L, Akiyama Y. 2013. Biochemical and structural insights into intramembrane metalloprotease mechanisms. Biochim Biophys Acta 1828:2873–2885. doi: 10.1016/j.bbamem.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrell D, Zhang Y, Olenic S, Kroos L. 2017. Bacillus subtilis intramembrane protease RasP activity in Escherichia coli and in vitro. J Bacteriol 199:e00381-17. doi: 10.1128/JB.00381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt TR, Scott EJ, Dyer DW. 2011. Whole-genome phylogenies of the family Bacillaceae and expansion of the sigma factor gene family in the Bacillus cereus species-group. BMC Genomics 12:430. doi: 10.1186/1471-2164-12-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinrich J, Wiegert T. 2006. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol Microbiol 62:566–579. doi: 10.1111/j.1365-2958.2006.05391.x. [DOI] [PubMed] [Google Scholar]
- 47.Castro AN, Lewerke LT, Hastie JL, Ellermeier CD. 2018. Signal peptidase is necessary and sufficient for site-1 cleavage of RsiV in Bacillus subtilis in response to lysozyme. J Bacteriol 200:e00663-17. doi: 10.1128/JB.00663-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, Gross CA. 2011. Signal integration by DegS and RseB governs the sigmaE-mediated envelope stress response in Escherichia coli. Proc Natl Acad Sci U S A 108:2106. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kocaoglu O, Tsui HCT, Winkler ME, Carlson EE. 2015. Profiling of β-lactam selectivity for penicillin-binding proteins in Streptococcus pneumoniae D39. Antimicrob Agents Chemother 59:3548–3555. doi: 10.1128/AAC.05142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kocaoglu O, Carlson EE. 2015. Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob Agents Chemother 59:2785–2790. doi: 10.1128/AAC.04552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. 2012. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol 7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilcks A, Jayaswal N, Lereclus D, Andrupl L. 1995. Characterization of plasmid pAW63, a second self-transmissible plasmid in Bacillus thuringiensis subsp. kurstaki HD73. Plasmid 73:1263–1270. doi: 10.1099/00221287-144-5-1263. [DOI] [PubMed] [Google Scholar]
- 53.Hoffmaster AR, Koehler TM. 1999. Autogenous regulation of the Bacillus anthracis pag operon. J Bacteriol 181:4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koehler TM. 2009. Bacillus anthracis physiology and genetics. Mol Aspects Med 30:386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 56.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods 77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bone EJ, Ellar DJ. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett 58:171–177. doi: 10.1016/0378-1097(89)90033-5. [DOI] [PubMed] [Google Scholar]
- 59.Lereclus D, Arantes O, Chaufaux J, Lecadet M-M. 1989. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett 60:211–217. doi: 10.1111/j.1574-6968.1989.tb03448.x. [DOI] [PubMed] [Google Scholar]
- 60.Griffith KL, Wolf RE. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun 290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- 61.Slauch JM, Silhavy TJ. 1991. Cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol 173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marini P, Li SJ, Gardiol D, Cronan JE, De Mendoza D. 1995. The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilis acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. J Bacteriol 177:7003–7006. doi: 10.1128/jb.177.23.7003-7006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karp PD, Billington R, Caspi R, Fulcher CA, Latendresse M, Kothari A, Keseler IM, Krummenacker M, Midford PE, Ong Q, Ong WK, Paley SM, Subhraveti P. 2017. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform 2017:bbx085. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ransom EM, Ellermeier CD, Weiss DS. 2015. Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl Environ Microbiol 81:1652–1660. doi: 10.1128/AEM.03446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP-RsiP is functional. B. thuringiensis rsiP1–80 (THE2628) containing tetracycline-inducible gfp-rsiP (EBT587) or empty vector (EBT561) was plated on LB–X-Gal without or with ATc (50 ng/ml). Download FIG S1, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GFP-RsiP localizes to the membrane. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360), tetracycline-inducible gfp-rsiP1–72 (EBT533), or empty vector (EBT169) was subcultured 1:50 with ATc (100 ng/ml) and grown at 30°C to late log phase. Two microliters was spotted on a 1% agarose pad for immobilization and imaged for GFP localization. Phase-contrast and fluorescence micrographs were recorded on an Olympus BX60 microscope with a 100 UPlanApo objective (numerical aperture, 1.35). For the GFP micrographs, a filter set from Chroma Technology Corp (catalog no. 41017) was used. The GFP filter consists of a 450- to 490-nm excitation filter, a 495-nm dichroic mirror (long pass), and a 500- to 550-nm emission filter. Micrographs were captured with a Hamamatsu Orca Flash 4.0 V2 complementary metal oxide semiconductor (CMOS) camera. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RsiP levels decrease in the presence of cefoxitin. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360) or empty vector (EBT169) was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cells were incubated with 5 μg/ml of cefoxitin for various times (0, 15, 30, 60, 120, or 180 min) (A) or increasing concentrations of cefoxitin (0, 0.05, 0.5, 5, 50, or 500 μg/ml) for 1 h (B). The immunoblot was probed with antisera against RsiP (α-RsiP76–275) followed by goat anti-rabbit IgG IR800CW (green). EV is wild type with pAH9 (EBT169), and GFP is wild type with pAH13 (UM20). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). Download FIG S3, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RsiP levels decrease in the presence of cefoxitin. B. thuringiensis expressing tetracycline-inducible gfp-rsiP (EBT360), empty vector (EBT169), or GFP alone (UM20) was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cells were incubated with increasing concentrations of cefoxitin (0, 0.05, 0.5, 5, 50, and 500 μg/ml) for 1 h. The immunoblot was probed with antisera against GFP (α-GFP) followed by goat anti-rabbit IgG IR800CW (green). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). The Western blot is shown in black and white (A) or color (B). Download FIG S4, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Amino acid alignment of RasP. An alignment of B. subtilis RasP and B. thuringiensis HD73_4301. The active site is marked by a red box. Download FIG S5, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RsiP degradation is dependent upon the site-2 protease RasP. B. thuringiensis containing a tetracycline-inducible copy of gfp-rsiP, wild type (EBT360) or ΔrasP (EBT366), was subcultured 1:50 into LB supplemented with ATc (50 ng/ml). At mid-log phase, cultures were incubated for 1 h without (−) or with (+) cefoxitin treatment (5 μg/ml) at 37°C. The immunoblot was probed with antisera against GFP (anti-GFP) followed by goat anti-rabbit IgG IR800CW (green). EV is wild type with pAH9 (EBT169), and GFP is wild type with pAH13 (UM20). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). Download FIG S6, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RsiP mutants result in constitutive σP activity. Alignment of RsiP mutants that result in nonsense, frameshift, or point mutations. Amino acid residues in red are indicative of the change in amino acid sequence due to mutations. The mutation rsiP1–232 was isolated 3 independent times while rsiP1–61 and rsiP1–15 were isolated twice each. The red box indicates the predicted transmembrane domain. Download FIG S7, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of RsiP mutants. B. thuringiensis containing either empty vector (wild type, EBT169; rsiP1–220, EBT564; rsiP1–80, EBT565; rsiP1–61, EBT566; rsiP1–16, EBT567) or PsigP-sigP-rsiP (wild type, EBT168; rsiP1–220, EBT560; rsiP1–80, EBT651; rsiP1–61, EBT562; rsiP1–16, EBT563) was grown overnight and spotted onto LB–X-Gal (100 μg/ml) plates lacking cefoxitin (A) or containing cefoxitin (5 μg/ml) (B). Download FIG S8, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Truncation of RsiP results in constitutive degradation in a RasP-dependent manner. B. thuringiensis containing a tetracycline-inducible copy of gfp-rsiP, gfp-rsiP1–72 (rsiP without the extracellular domain) or gfp-rsiP1–53 (rsiP without the transmembrane and extracellular domains), was constructed in either wild type or a ΔrasP mutant strain (GFP-RsiP wild type [rasP+], EBT360; GFP-RsiP ΔrasP, EBT366; GFP-RsiP1–53 wild type [rasP+], EBT518; GFP-RsiP1–53 ΔrasP, EBT510; GFP-RsiP1–72 wild type [rasP+], EBT516; GFP-RsiP1–72 ΔrasP, EBT533). Strains were subcultured 1:50 into LB supplemented with ATc (100 ng/ml) and at mid-log phase were incubated for 2 h without (−) or with (+) cefoxitin treatment (5 μg/ml) at 37°C. The immunoblot was probed with antisera against GFP (anti-GFP) followed by goat anti-rabbit IgG IR800CW (green). Streptavidin IR680LT (red) was used to detect HD73_4231 (PycA homolog), which served as a loading control (62, 63). Download FIG S9, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides. Download Table S1, PDF file, 0.06 MB (65.5KB, pdf) .
Copyright © 2019 Ho et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.