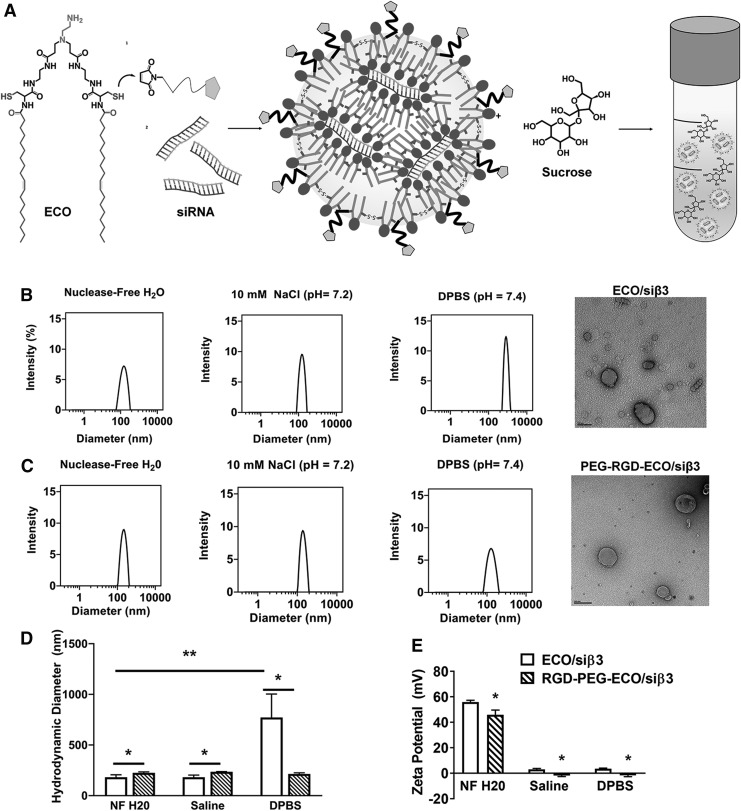
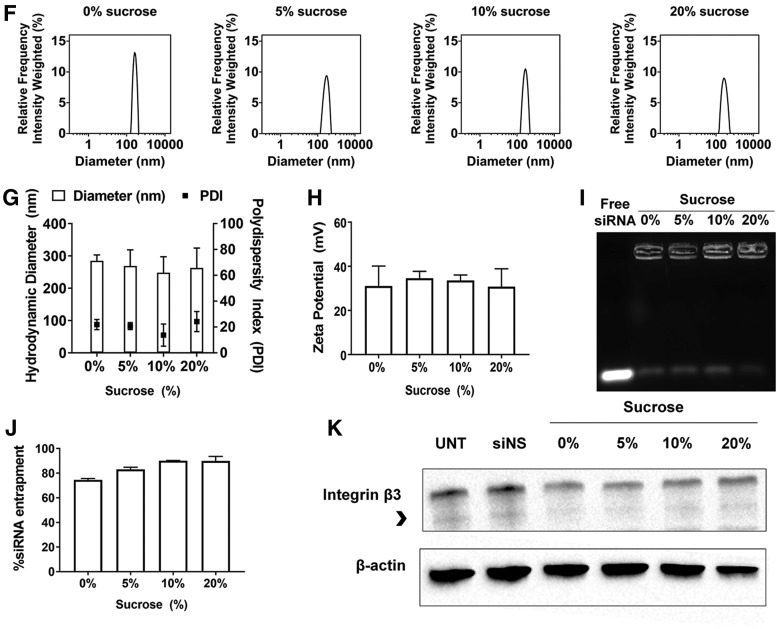
FIG. 1.
Characterization of freshly prepared ECO/siβ3 nanoparticles with and without MAL-PEG-RGD in aqueous solutions and in the presence or absence of sucrose. (A) Schematic of experimental design. Representative intensity peaks of ECO/siβ3 nanoparticles (B) and RGD-PEG-ECO/siβ3 nanoparticles (C) in nuclease-free water, 10 mM NaCl (pH = 7.2), and DPBS (CaCl2: 0.9 mM, MgCl2: 0.5 mM, KCl: 2.67 mM, KH2PO4: 1.47 mM, NaCl: 137.9 mM, Na2HPO4: 8 mM). Transmission electron microscopy images of ECO/siβ3 nanoparticles (B) and RGD-PEG-ECO/siβ3 nanoparticles (C). Comparison of the hydrodynamic diameters (D) and zeta potential (E) of ECO/siβ3 nanoparticles and RGD-PEG-ECO/siβ3 nanoparticles in aqueous solutions. (F) Representative intensity peaks of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose in nuclease-free water. Hydrodynamic diameter, polydispersity index (G), and zeta potential (H) of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose in nuclease-free water. (I) Agarose gel retardation of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose compared to free siβ3. (J) RiboGreen assay quantifying siRNA entrapment of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose. (K) Western blot analysis of β3 integrin expression (indicated by the arrowhead) in BT549 cells 48 h after RGD-PEG-ECO/siβ3 nanoparticle treatment (error bars denote SEM, *P < 0.05, **P < 0.01). DPBS, Dulbecco's phosphate-buffered saline; MAL, maleimide; PEG, polyethylene glycol; SEM, standard error of the mean.