Abstract
Genome sequencing of environmental bacteria allows identification of biosynthetic gene clusters encoding unusual combinations of enzymes that produce unknown natural products. We identified a pathway in which a ribosomally synthesized small peptide serves as a scaffold for non-ribosomal peptide extension and chemical modification. Amino acids are transferred to the C-terminus of the peptide through ATP and amino acyl-tRNA-dependent chemistry that is independent of the ribosome. Oxidative rearrangement, carboxymethylation, and proteolysis of a terminal cysteine yields an amino acid derived small-molecule. Microcrystal electron diffraction demonstrates that the resulting product is isosteric to glutamate. We show that a similar peptide extension is used during the biosynthesis of the ammosamides, cytotoxic pyrroloquinoline alkaloids. These results suggest an alternative paradigm for biosynthesis of amino acid derived natural products.
One Sentence Summary:
Amino acids are added to the C-terminus of gene-encoded scaffold peptides, followed by maturation and proteolytic release.
Bacteria produce many small-molecule natural products that play important roles in communication, symbiosis, and competition (1). Historically these compounds have been discovered by activity-based screens, but an alternative avenue for their discovery starts with identification of their biosynthetic gene clusters now that bacterial genomes have revealed the tremendous diversity of natural products that remain to be discovered (2). In this study we focus on a group of biosynthetic gene clusters for which the final products were not known and could not be predicted.
Ribosomally-synthesized and posttranslationally-modified peptides (RiPPs) (3) include lantibiotics and thiopeptides that are used in food and agriculture (4). They are biosynthesized from a precursor peptide consisting of a leader peptide that serves as a recognition motif for the biosynthetic enzymes and a core peptide that is converted to the final product. During their maturation, Ser and Thr residues are glutamylated by LanB enzymes in a glutamyl-tRNA dependent mechanism (5, 6). Subsequently, the glutamate is eliminated to generate dehydroamino acids (Fig. 1A). A survey of >100,000 publicly available bacterial genomes revealed more than 600 genes encoding LanB-like proteins in which the elimination domain is not present within the cluster or genome.
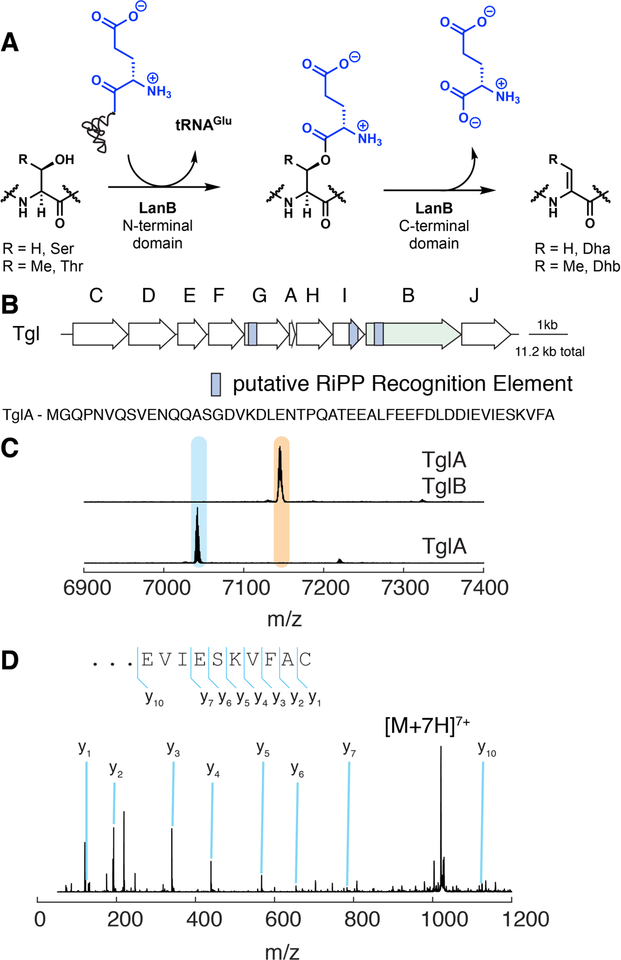
Figure 1. Function of a small LanB enzyme, TglB, found in P. syringae.
(A) LanB enzymes glutamylate Ser/Thr residues and subsequently eliminate the glutamate to form dehydroamino acids. Small LanB proteins lack the elimination domain. Dha, dehydroalanine; Dhb, dehydrobutyrine. (B) Biosynthetic gene cluster in P. syringae encoding a small LanB. (C) Matrix-assisted laser desorption ionization with time-of-flight (MALDI-TOF) mass spectra of TglA coexpressed with TglB. (D) Analysis of the TglB product by tandem electrospray ionization (ESI) mass spectrometry.
In the plant pathogen Pseudomonas syringae pv. maculicola ES4326, such a protein (TglB) is encoded near an open reading frame encoding a 50-amino acid peptide (TglA; Fig. 1B). Co-expression of His6-TglA and TglB in Escherichia coli and subsequent purification of the peptide demonstrated an increase in mass by 103 Da (Fig. 1C). This increase is inconsistent with glutamylation, but could be the result of condensation with a cysteine residue. High resolution tandem mass spectrometry analysis of the peptide suggested that the adduct was attached to the C-terminal alanine instead of the anticipated ester linkage to a Ser in the peptide (Fig. 1D). We expressed TglA and TglB individually as His6-tagged proteins and purified them. In vitro incubation with Cys, ATP, tRNACys and Cys tRNA synthetase (CysRS) resulted in the same product (TglA-Cys; Fig. 2A) as that isolated from co-expression in E. coli confirming that TglB adds a Cys to the C-terminus of TglA in a tRNA dependent manner (fig. S1A). This C-terminal peptide extension not only constitutes a previously unknown posttranslational modification but also seems counterintuitive since a more logical route to the product appears to be encoding the Cys on tglA. We next purified Cys-tRNACys and showed that TglB does not transfer the Cys to the C-terminus of TglA unless ATP is present, which is converted to ADP and phosphate (fig. S1B). Performing the reaction in buffer made with H2 18O and subsequent MS analysis demonstrated that the product contains one 18O atom (fig. S1C), and addition of hydroxyl amine to the assay allowed trapping of C-terminally activated TglA as the hydroxamate (fig. S1D). These findings are consistent with activation of the C-terminus of TglA by phosphorylation, subsequent amide bond formation with the amino group of Cys-tRNA, and release of the tRNA by hydrolysis (fig. S1E). The observations rule out the use of the activated ester of Cys-tRNA for the non-ribosomal peptide extension (fig. S1E). TglB accepted a 12-mer peptide corresponding to the C-terminus of TglA as a minimal substrate (fig. S1F), and kinetic experiments showed that TglB has a turnover number of 28 min−1 using full length TglA (fig. S1G).
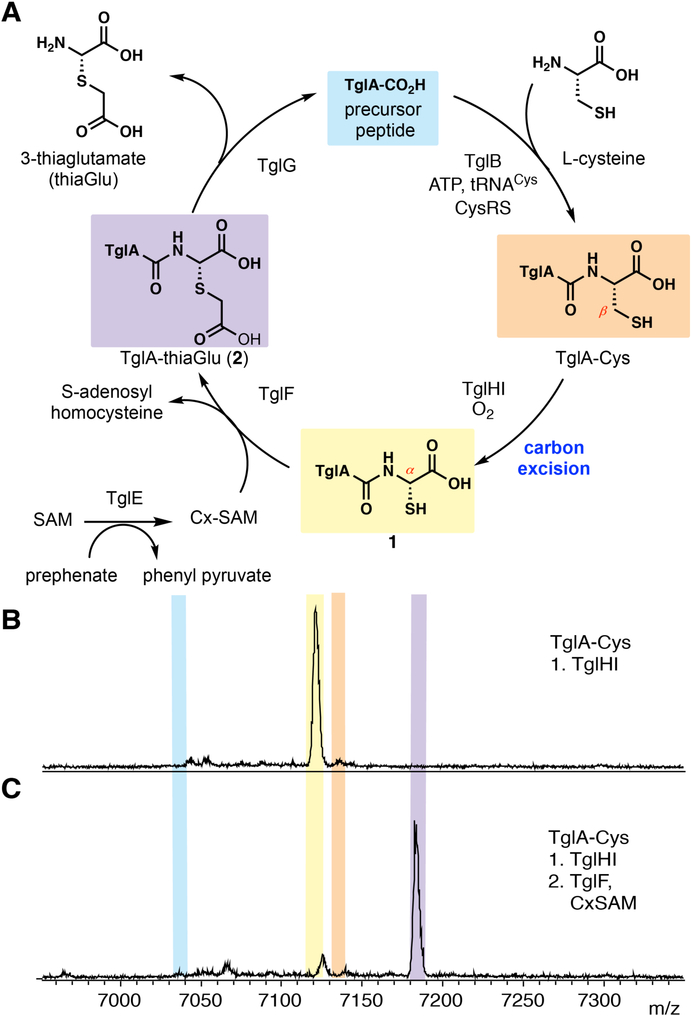
Figure 2. The cysteine added by TglB is modified by other enzymes from the tgl cluster.
(A) Inferred biosynthetic pathway towards 3–thiaglutamate. (B) MALDI-TOF mass spectrum of in vitro reaction of TglHI with TglA-Cys. (C) MALDI-TOF mass spectrum of in vitro reaction of TglF with compound 1. Color-coding of shaded peaks in panels B and C are shown in panel A.
We next interrogated the other proteins encoded in the biosynthetic gene cluster. TglH has low homology to a structurally characterized dinuclear non-heme iron dependent protein for which no activity has been reported (7). The C-terminal domain of TglI has homology with known leader peptide binding domains in RiPP biosynthetic enzymes (Fig. 1B) (5, 8). We co-expressed TglA with TglB, TglH and TglI in E. coli and isolated a product that was decreased in mass by 14 Da from TglA-Cys (fig. S2A). We treated the peptide with trypsin to generate a C-terminal tetrapeptide. Chemical assays with thiol- and carboxylate-reactive electrophiles indicated that the product still contained these functional groups (fig. S3) suggesting structure 1 as the product of TglHI (Fig. 2A). We next repeated this experiment but using an E. coli strain that is auxotrophic for Cys and that was grown in minimal media supplemented with 13C-labeled Cys. Isolation of the peptide and analysis by MS showed that it is the cysteine β-carbon that is removed (fig. S4).
The biosynthetic cluster also contains a pair of genes (tglEF) encoding proteins similar to a recently characterized carboxy-S-adenosylmethionine (Cx-SAM) synthase and a SAM-dependent methyltransferase, respectively (9, 10). We added compound 1 to Cx-SAM and TglF in vitro and isolated product 2 with a mass increase of 58 Da (Fig. 2C), consistent with carboxymethylation of a thiol. This hypothesis was confirmed by treating the TglHI product with iodoacetic acid, which resulted in the same outcome, as did co-expression of TglABEFHI in E. coli (fig. S2B). The in vitro prepared peptide was treated with trypsin and the C-terminal tetrapeptide 3 was characterized by 1H NMR spectroscopy and tandem MS, which supported structure 2 for the TglF product (Fig. 2A; fig. S5). Given the unusual architecture, we also chemically synthesized peptide 3 as two diastereomers (Supplementary Information) and demonstrated that the 1H NMR spectrum of one isomer was identical to the enzymatic product (fig. S5). We tried to obtain crystals to assign the stereochemistry of either isomer and made several chemical derivatives but were unable to obtain crystals for X-ray diffraction.
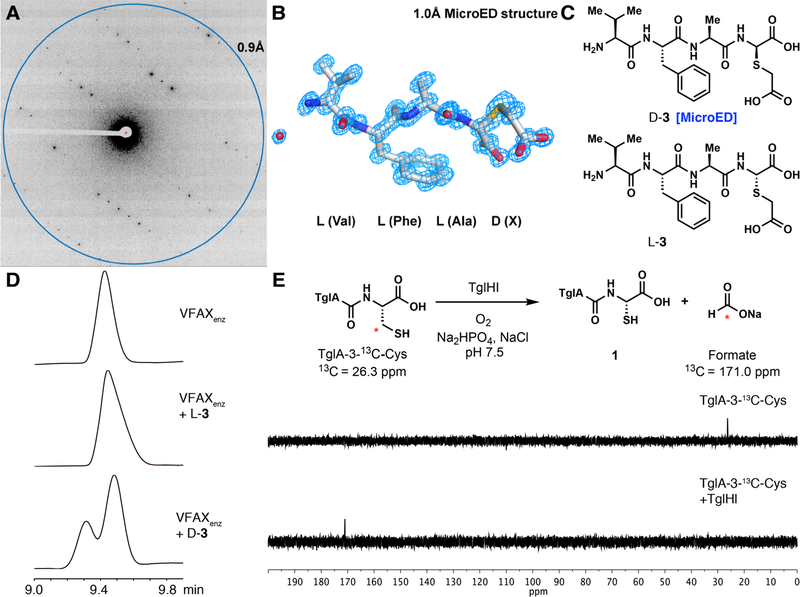
We next turned to the cryoEM method microcrystal electron diffraction (MicroED) (11–13). A small amount of powder of the diastereomer that was obtained in higher amounts and in more pure form was placed onto an EM grid, plunged into liquid nitrogen and investigated under cryogenic conditions in an electron microscope. The seemingly amorphous powder contained numerous nanocrystals on the grid suitable for MicroED analysis, each consisting roughly of femtograms of material that diffracted to ~1 Å resolution. MicroED data was collected from each nanocrystal but the sample was highly susceptible to beam damage such that no useful diffraction was observed after the first few frames of the MicroED movie. Despite >150 data sets collected on a CMOS-based CetaD camera, nanocrystals succumbed to radiation damage too fast preventing structure determination. It is possible that the peptide was particularly susceptible to damage because of the 3-thiaglutamate, consistent with an earlier study that showed that radiation damage is particularly prevalent at Cys residues (14). We then turned to the Falcon III direct electron detector, one of the most sensitive cameras for cryoEM that was recently demonstrated to be suitable for MicroED data collection and structure determination and that minimizes radiation damage because of its high sensitivity and high frame rate (15). Atomic resolution data from seven nanocrystals were collected each covering an angular range of ~50° before damage was observed. Data from five nanocrystals were merged to yield a 96% complete data set to 1.0 Å resolution and the structure was determined by direct methods (Fig. 3; Crystallographic Table S3; Supplementary Information). The atomic-resolution MicroED structure was thus solved demonstrating the D configuration of the 3-thiaGlu in this peptide (D-3), which in turn provided the stereochemical assignment for L-3 which co-elutes with and has the same spectral data as the enzymatic product. These results demonstrate that the TglHI-catalyzed reaction occurred with retention of configuration at the α-carbon (Fig. 3B–D). These findings highlight the utility of MicroED to determine the structure and stereochemistry of a previously unknown natural product. Thus, collectively TglBEFHI convert TglA into a peptide containing L-3-thiaglutamate at its C-terminus (TglA-thiaGlu, 2; Fig. 2A).
Figure 3. In vitro TglHI reacts with 13C-labeled TglA-Cys to produce 13C-formate and 1 with retention of configuration.
(A) Diffraction pattern of D-3 with resolution ring at 0.9 Å. (B) Atomic MicroED structure of D-3 determined at 1.0 Å resolution. X refers to 3-thiaGlu. (C) Structure of chemically synthesized tetrapeptides (VFAX) containing D-thiaGlu (D-3) and L-thiaGlu (L-3). (D) Determination of stereochemical configuration of thiaGlu by comparison with synthetic standards. High-performance liquid chromatograms are shown. VFAXenz was obtained by TglHI modification of TglA-Cys followed by 2-iodoacetic acid alkylation and trypsin digest. (E) 13C NMR spectra showing the β-carbon of the C-terminal cysteine in 13C-labeled TglA-Cys (26.3 ppm, top), and a new signal at 171.0 ppm that corresponds to 13C-formate after reaction with TglHI (bottom).
We next investigated the TglHI-catalyzed reaction with purified proteins. Neither protein could be expressed in soluble form individually, but co-expression resulted in co-purification and metal analysis indicated TglHI contained 2.5 Fe. In vitro TglHI converted TglA-Cys to 1 under aerobic conditions with a turnover number of 1.1 min−1 (Fig. 2B) whereas under low oxygen concentrations product formation was negligible, confirming oxygen-dependency of the reaction (fig. S6A). To investigate if TglHI can functionalize internal cysteine residues, the extension mutant TglA-CysAla was prepared. This peptide was not modified by TglHI (fig. S6B). TglHI also did not modify other unrelated peptides that end in Cys (fig. S6C), and N-terminal truncation of TglA-Cys led to diminished or abolished TglHI activity (fig. S6D). Thus, the enzyme has high specificity for TglA-Cys. To identify the fate of the lost carbon atom, 13C-labeled TglA-Cys was reacted with TglHI and formate was observed by 13C NMR spectroscopy (Fig. 3E). Moreover, when [2,3,3,−2H]-Cys was used, the product contained one deuterium illustrating that the α-hydrogen is likely not removed during the transformation (fig. S4D). Thus, TglHI catalyzes a net four-electron oxidation of TglA-Cys, modifying the redox states of both the α and β carbons of the C-terminal cysteine installed by TglB. Based on the in vitro studies, we propose a mechanism for the formation of 1 and formate from TglA-Cys (fig. S7). The chemistry catalyzed by TglHI expands the range of post-translational modifications in natural product biosynthesis (16) to include a remarkable excision of a methylene group from cysteine. Additional TglHI-like enzymes are present in the genomes (fig. S8) including in the biosynthetic gene cluster encoding the methanobactin precursor (17–19).
The last four genes in the biosynthetic cluster encode a putative membrane bound protease (TglG), a putative pyridoxal-phosphate dependent enzyme (TglC) that is sometimes missing in homologous clusters, and two putative transporters (TglD and TglJ). Like TglB and TglI, TglG contains a RiPP leader peptide recognition motif suggesting it will act on a TglA-derived peptide (Fig. 1B) and homologous enzymes have cytoplasmic active sites (20). When TglA-thiaGlu was exposed to the membrane fraction of cell lysate of E. coli expressing GFP-TglG, the peptide was cleanly converted into TglA (fig. S9). TglA-Glu was also a substrate but not TglA-GluAla, illustrating that the protease cannot distinguish Glu and 3-thiaGlu but does not tolerate extension of the peptide. Thus, TglA appears to be a scaffold on which 3-thiaGlu is assembled and final proteolytic release regenerates TglA for another round of biosynthesis (Fig. 2A). Were cysteine merely encoded in tglA, then each ribosomally produced peptide could make only a single 3-thiaglutamate. Instead, the use of TglA as a scaffold peptide is conceptually more efficient than the stoichiometric use of leader peptide in other RiPP pathways (4). At present we do not know the function of 3-thiaGlu, nor whether this unstable compound is further chemically modified. Plants were recently shown to use Glu for a systemic signaling response to pathogens (21), and it is possible that 3-thiaGlu or a product derived from it interferes with Glu signaling similarly to other anti-metabolite toxins made by P. syringae that block jasmonate and ethylene signaling pathways (22).
We note that 3-thiaGlu is not a RiPP because it is not ribosomally synthesized, but it is made by posttranslational modification reactions. Perhaps this unusual pathway evolved because of the significant relative burden of leader peptide production for a single amino acid product. Bioinformatic prediction of TglA transcriptional regulation (23) suggests precursor production is not driven by a separate promoter, which is consistent with putative catalytic use of the peptide (fig. S10). This contrasts with most RiPP pathways in which expression of the substrate peptide is controlled by its own promoter followed by a read-through transcriptional terminator to allow the precursor peptide to be present in excess over the biosynthetic machinery (24, 25).
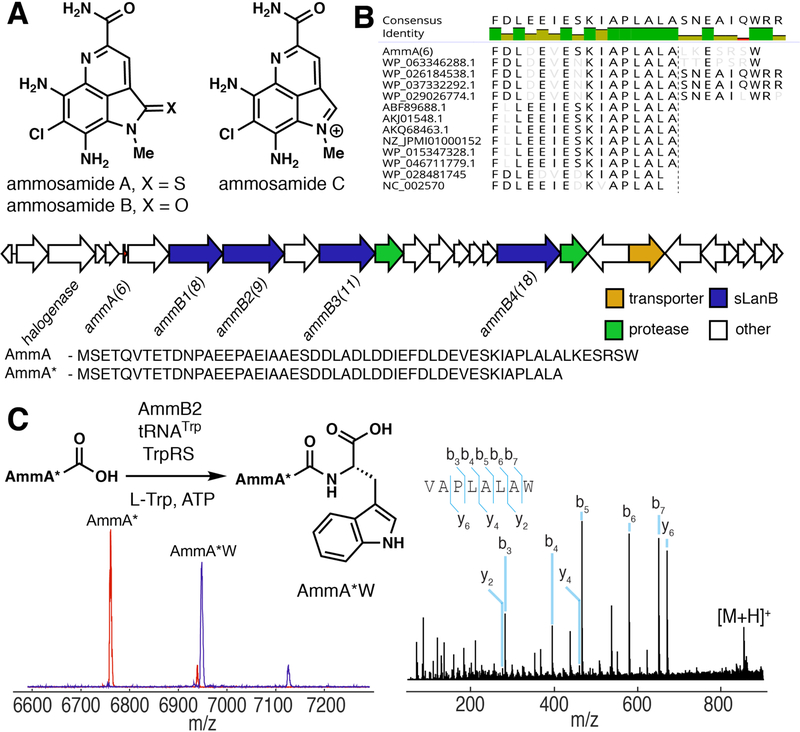
It is the Cys-tRNA-dependent enzyme TglB that allows the proposed catalytic use of TglA. Similar small LanB-encoding genes are found in several bacterial phyla, with some clusters encoding multiple such proteins and a range of additional putative modification enzymes (fig. S11). To assess the generality of the function of small LanB proteins and provide further support for a catalytic role of the scaffold peptide, we investigated ammosamide biosynthesis. A previous study of these Trp-derived pyrroloquinoline natural products (Fig. 4A) hinted that the compounds could be derived from a small peptide AmmA ending in Trp encoded in the gene cluster (Fig. 4B) (26). However, when this Trp was mutated to Ser or deleted altogether, ammosamide was still produced (26). The ammosamide gene cluster encodes four small LanB proteins. We tested all four for activity in vitro and in E. coli with AmmA (previously annotated Amm6) and AmmA lacking the C-terminal Trp but observed no activity. We noted that AmmA has homology with other peptides encoded in clusters with small LanB proteins (Fig. 4B), but that AmmA appears to have a C-terminal extension. When we removed this extension, AmmB2 (previously annotated Amm9), but not the other three AmmB proteins, added a Trp in a Trp-tRNA dependent fashion to the C-terminus of the peptide in vitro and in E. coli (Fig. 4C). This finding explains the observation that mutation or deletion of the C-terminal Trp still resulted in ammosamide production, and supports catalytic use of the peptide. Such use provides an attractive explanation for the 134 mg/L of ammosamide C produced by the producing bacterium (26) because stoichiometric use would require production of 3.0 g of AmmA. Given this second example of tRNA-dependent activity, we suggest the name peptide-amino acyl tRNA ligase (PEARL) for the small LanB proteins. The biosynthesis of a metabolite on a small peptide scaffold is uncommon, with the closest similarity found in the biosynthesis of amino acids linked by isopeptide bonds to a glutamate residue on amino-carrier proteins in some bacteria (27, 28).
Figure 4. Ammosamide biosynthesis involves addition of L-Trp to the C-terminus of a ribosomally synthesized peptide.
(A) Pyrolloquinoline alkaloids ammosamides A-C. (B) Sequence alignment of the C-terminus of the AmmA precursor peptide and its homologs showing a C-terminal extension for AmmA relative to most homologs. The gene cluster for ammosamide biosynthesis in Streptomyces sp. CNR698 comprises 27 orfs. The encoded proteins include four small LanBs, two proteases, one halogenase and a transporter. (C) AmmB2 adds L-Trp to AmmA*, a truncated peptide of AmmA, to afford AmmA*W in vitro in an ATP, tRNATrp and Trp-RS dependent reaction. Red MALDI-TOF mass spectrum is AmmA* and the blue spectrum shows the product of the reaction. HR-ESI MS/MS confirms addition of L-Trp to the C-terminus.
Supplementary Material
Acknowledgments
P. syringae pv. maculicola ES4326 was provided by Prof. Darrell Desveaux (University of Toronto). We thank Rebecca Splain (van der Donk lab) and Michael W. Martynowycz and Johan Hattne (Gonen lab, UCLA) for advice and useful discussions. Funding: This work was supported by the National Institutes of Health (R37 GM058822 to W.A.V.; F32 GM129944 to C.P.T.; F32 GM120868 to M.A.F). T.G. and W.A.V. are Investigators of the Howard Hughes Medical Institute.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: The authors declare no competing financial interests. M.A.F. is employed by Science and his editorial access to the paper was blocked.
Data and materials availability: All data supporting the findings in this study are provided in the main text and supplementary materials.
References
- 1.Adnani N, Rajski SR, Bugni TS, Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep 34, 784–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cimermancic P et al. , Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortega MA, van der Donk WA, New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol 23, 31–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnison PG et al. , Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega MA et al. , Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson GA, Zhang Z, Tietz JI, Mitchell DA, van der Donk WA, In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J. Am. Chem. Soc 137, 16012–16015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crystal structure of a duf692 family protein (hs_1138) from haemophilus somnus 129pt at 2.20 A resolution. PDB 3BWW, DOI 10.2210/pdb2213BWW/pdb (2008). [DOI] [Google Scholar]
- 8.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA, A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol 11, 564–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J et al. , Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 498, 123–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J et al. , Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification. Nucleic Acids Res. 43, 4602–4613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CG et al. , The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination. ACS Cent. Sci 4, 1587–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nannenga BL, Shi D, Leslie AGW, Gonen T, High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods 11, 927–930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi D, Nannenga BL, Iadanza MG, Gonen T, Three-dimensional electron crystallography of protein microcrystals. eLife 2, e01345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattne J et al. , Analysis of Global and Site-Specific Radiation Damage in Cryo-EM. Structure 26, 759–766.e754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattne J, Martynowycz M, Gonen T, MicroED with the Falcon III direct electron detector. BioRxiv, doi: 10.1101/615484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntosh JA, Donia MS, Schmidt EW, Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep 26, 537–559 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney GE, Rosenzweig AC, Chemistry and biology of the copper chelator methanobactin. ACS Chem. Biol 7, 260–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HJ et al. , Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305, 1612–1615 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Kenney GE et al. , The biosynthesis of methanobactin. Science 359, 1411–1416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng L et al. , Structure of a site-2 protease family intramembrane metalloprotease. Science 318, 1608–1612 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Toyota M et al. , Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–1115 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Arrebola E, Cazorla FM, Perez-Garcia A, de Vicente A, Chemical and metabolic aspects of antimetabolite toxins produced by Pseudomonas syringae pathovars. Toxins 3, 1089–1110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MN, Alm EJ, Arkin AP, Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res. 33, 3224–3234 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee C, Paul M, Xie L, van der Donk WA, Biosynthesis and mode of action of lantibiotics. Chem. Rev 105, 633–684 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Dufour A, Hindré T, Haras D, Le Pennec JP, The biology of the lantibiotics of the lacticin 481 subgroup is coming of age. FEMS Microbiol. Rev 31, 134–167 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Jordan PA, Moore BS, Biosynthetic Pathway Connects Cryptic Ribosomally Synthesized Posttranslationally Modified Peptide Genes with Pyrroloquinoline Alkaloids. Cell Chem. Biol 23, 1504–1514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasebe F et al. , Amino-group carrier-protein-mediated secondary metabolite biosynthesis in Streptomyces. Nat. Chem. Biol 12, 967–972 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Ouchi T et al. , Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus. Nat. Chem. Biol 9, 277–283 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Sullivan MJ, Petty NK, Beatson SA, Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung CT, Niemela SL, Miller RH, One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A 86, 2172–2175 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thibodeaux CJ, Ha T, van der Donk WA, A price to pay for relaxed substrate specificity: a comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J. Am. Chem. Soc 136, 17513–17529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennessy DJ, Reid GR, Smith FE, Thompson SL, Ferene — a new spectrophotometric reagent for iron. Can. J. Chem 62, 721–724 (1984). [Google Scholar]
- 33.Zhu S et al. , Insights into the unique phosphorylation of the lasso peptide paeninodin. J. Biol. Chem 291, 13662–13678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogh A, Larsson B, von Heijne G, Sonnhammer EL, Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Sonnhammer EL, von Heijne G, Krogh A, A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol 6, 175–182 (1998). [PubMed] [Google Scholar]
- 36.Solovyev V, Salamov A, in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, Li RW, Ed. (Nova Science Publishers, New York, 2010), pp. 61–78. [Google Scholar]
- 37.Kabsch W, Xds. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W, Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheldrick GM, Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem 71, 3–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheldrick GM, SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr. A Found. Adv 71, 3–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samant MP et al. , Structure-activity relationship studies of gonadotropin-releasing hormone antagonists containing S-aryl/alkyl norcysteines and their oxidized derivatives. J. Med. Chem 50, 2067–2077 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dardonville C et al. , Selective Inhibition of Trypanosoma brucei 6-Phosphogluconate Dehydrogenase by High-Energy Intermediate and Transition-State Analogues. J. Med. Chem 47, 3427–3437 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Gisin BF, The Preparation of Merrifield-Resins Through Total Esterification With Cesium Salts. Helv. Chim. Acta 56, 1476–1482 (1973). [Google Scholar]
- 44.Brown PM et al. , Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism. Proc. Natl. Acad. Sci. U.S.A 103, 15032–15037 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xing G et al. , Evidence for C-H cleavage by an iron-superoxide complex in the glycol cleavage reaction catalyzed by myo-inositol oxygenase. Proc. Natl. Acad. Sci. U.S.A 103, 6130–6135 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peck SC et al. , O-H Activation by an Unexpected Ferryl Intermediate during Catalysis by 2-Hydroxyethylphosphonate Dioxygenase. J. Am. Chem. Soc 139, 2045–2052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamanaha E et al. , Spectroscopic Evidence for the Two C-H-Cleaving Intermediates of Aspergillus nidulans Isopenicillin N Synthase. J. Am. Chem. Soc 138, 8862–8874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerlt JA et al. , Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solovyev VV, Shahmuradov IA, Salamov AA, Identification of promoter regions and regulatory sites. Methods Mol. Biol 674, 57–83 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.