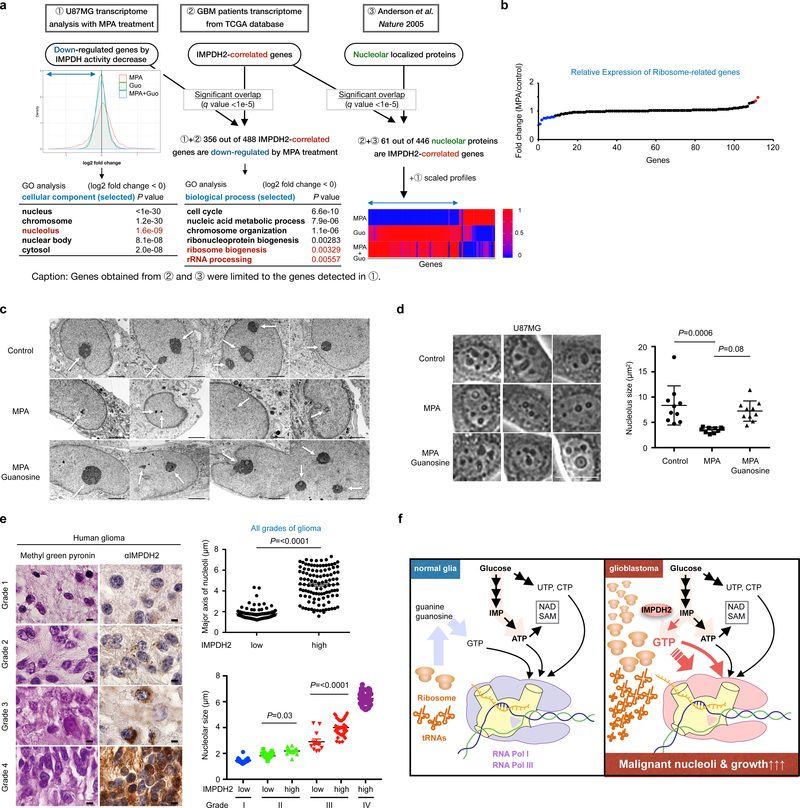
Fig. 7 |. IMPDH2 upregulation is critical for nucleolar transformation in GBM.
a, Two-dimensional (2D) transcriptome screening reveals IMPDH2-network and its association with nucleolar activity in GBM. The Gene Ontology (GO) analysis of IMPDH activity-associated genes (left) and the genes associated with both IMPDH2-expression and IMPDH-activity (right) are shown in the tables. Bottom right show the expression of genes encoding nucleolar localizing proteins that are correlated with IMPDH2 mRNA expression in the GBM patient transcriptome. b, 10 μM MPA treatment (24 h) marginally affect the expression of genes encoding ribosomal subunits in U87MG. Red color is for >1.3-fold increased genes, while blue for the >1.3-fold decreased genes. n=1 experiment.
c, U87MG cells were treated with or without 10 μM MPA, guanosine for 72 h and analyzed by transmission electron microscope (x 6,000). Scale bar indicates 2 μm. n=1 experiment.
d, IMPDH inhibition leads to decrease nucleolar size in U87MG. The area of nucleoli from 10 cells was assessed at 24 h of 10 μM MPA, 100 μM guanosine. Data are presented as mean±s.d. Average nucleolus sizes from 10 different cells in each group were indicated. The results were confirmed in two independent experiments. One-way ANOVA. Scale bar indicates 50 μm.
e, IMPDH2 upregulation and glioma malignancy are positively correlated with nucleolar hypertrophy in glioma patients (left). Methyl green pyronin and IMPDH2 staining on human glioma tissue array. Upper right: major axis of nucleoli in human glioma tissue array (all grades of glioma) according to IMPDH2 expression levels (low: n=115, high: n=109 specimens). Lower right: major axis of nucleoli in grade I to IV glioma specimens according to IMPDH2 expression levels (Grade I, low: n=21, Grade II, low: n=61, high: n=16, Grade III, low: n=12, high: n=41, Grade IV, high: n=41 specimens). Data are presented as mean±s.e.m., Mann-Whitney test (two-sided) (upper right), one-way ANOVA (lower right). Scale bar indicates 50 μm.
f, A schematic working model. Among four ribonucleotides, GTP biosynthesis from glucose is elevated in glioblastoma cells to fuel rRNA and tRNA via Pol I and Pol III transcription for their malignant growth. Less proliferative glial cells recycle ribonucleotides.