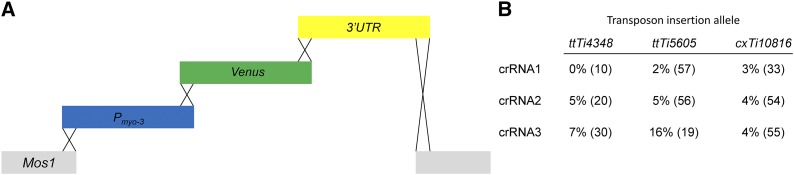
Figure 2.
Mos1 crRNA efficacy using recombineering with linear PCR fragments. A) Schematic (not to scale). A Cas9-mediated double strand break in the Mos1 transposon sequence (gray) was induced using crRNA1, crRNA2, or crRNA3. The break was repaired with three PCR amplicons to drive Venus expression in the body wall muscles. Crossed lines indicate homologous DNA sequences, which overlap by 35 bp. Pmyo-3 (blue, 2573 bp), Venus (Green, 810 bp) and unc-54 3′ UTR (yellow, 853 bp). B) The number of successfully injected P0 broods scored for crRNA1 was 3, 13, and 7 for ttTi4348, ttTi5605 and cxTi10816, respectively; crRNA2, 9, 13, 13; crRNA3, 5, 3, 10. The resulting dpy-10 progeny were screened for yellow fluorescent body wall muscles. Data are the percent integrations among the fluorescent dpy-10 edits. The number of fluorescent dpy-10 worms screened is in parentheses. The total F1 dpy-10 edits (non-fluorescent plus fluorescent) screened for crRNA1 was 30, 123, and 85 for ttTi4348, ttTi5605 and cxTi10816, respectively; crRNA2, 177, 199, 264; crRNA3, 74, 38, 141. PCR amplicons were injected at 0.147 - 0.210 pmol/µl in mixes containing crRNA1, 0.202 - 0.217 pmol/µl in mixes containing crRNA2, and 0.147 - 0.215 pmol/µl in mixes containing crRNA3. All PCR amplicons are listed in Table 3. PCR amplicons for crRNA1 are 2.1 (Pmyo-3), 2.2 (Venus), and 2.3 (3′ UTR) for crRNA1; 2.4 (Pmyo-3), 2.2 (Venus), and 2.5 (3′ UTR) for crRNA2; 2.6 (Pmyo-3), 2.2 (Venus), 2.7 (3′ UTR) for crRNA3.