Abstract
Introduction
Recent advances in genomic technology have allowed better delineation of renal conditions, the identification of new kidney disease genes and subsequent targets for therapy. To date, however, the utility of genomic testing in a clinically ascertained, prospectively recruited kidney disease cohort remains unknown. The aim of this study is to explore the clinical utility and cost-effectiveness of genomic testing within a national cohort of patients with suspected genetic kidney disease who attend multidisciplinary renal genetics clinics.
Methods and analysis
This is a prospective observational cohort study performed at 16 centres throughout Australia. Patients will be included if they are referred to one of the multidisciplinary renal genetics clinics and are deemed likely to have a genetic basis to their kidney disease by the multidisciplinary renal genetics team. The expected cohort consists of 360 adult and paediatric patients recruited by December 2018 with ongoing validation cohort of 140 patients who will be recruited until June 2020. The primary outcome will be the proportion of patients who receive a molecular diagnosis via genomic testing (diagnostic rate) compared with usual care. Secondary outcomes will include change in clinical diagnosis following genomic testing, change in clinical management following genomic testing and the cost-effectiveness of genomic testing compared with usual care.
Ethics and dissemination
The project has received ethics approval from the Melbourne Health Human Research Ethics Committee as part of the Australian Genomics Health Alliance protocol: HREC/16/MH/251. All participants will provide written informed consent for data collection and to undergo clinically relevant genetic/genomic testing. The results of this study will be published in peer-reviewed journals and will also be presented at national and international conferences.
Keywords: chronic renal failure, genetic kidney disease, genetics, genomics, nephrology
Strengths and limitations of this study.
This is a prospectively conducted multicentre national study in which patient data will be captured in a pragmatic clinical setting.
Patients will be recruited for clinically indicated genomic testing for suspected genetic kidney disorders, which may allow better generalisability of results.
Genomic testing is clinically indicated and performed routinely in some participating states/centres in Australia, and therefore it would be unethical to have a control arm for comparison as this would deny some patients of testing.
This study will contribute to the future research and clinical service redesign by establishing the utility of genomic testing in a kidney disease cohort from patient, clinician and health resource perspectives.
Introduction
Genetic kidney disease (GKD) accounts for 10% of adults with chronic kidney disease (CKD),1 with a monogenic cause being identified in around 20% of those with early-onset CKD.2 Recent advances in genomic sequencing have enabled rapid and cost-effective sequencing of large amounts of DNA3 via massively parallel sequencing, otherwise known as next generation sequencing (NGS). This, in turn, has led to better delineation of GKD, the identification of new renal disease genes and subsequent targets for therapy.4 Moreover, the clinical implementation of genomic testing has increased the number of patients receiving a timely and accurate genetic diagnosis.5 NGS-based genetic testing has demonstrated a monogenic cause in 20% of patients with early-onset CKD, and almost 10% in an unselected cohort of 3000 adults with CKD.2 6 A genomic diagnosis has many potential benefits including enabling targeted therapies,7–9 preventing the use of inappropriate treatments10 and reducing the use of invasive diagnostic investigations such as renal biopsy. In addition to concluding a sometimes protracted diagnostic odyssey, a genomic diagnosis may also provide prognostic information, inform targeted surveillance for extrarenal complications and facilitate transplantation and reproductive planning. Despite these potential benefits, there is a paucity of comprehensive evaluations of clinical utility and health economic impact of genomic testing in kidney disease cohorts.
Multifaceted novel approaches are required to address the complexities of successfully implementing genomic technologies into clinical care. Diagnostic renal genetics clinics (RGC) apply a multidisciplinary team approach through collaboration between adult and paediatric nephrologists, clinical geneticists, genetic counsellors and diagnostic laboratory scientists. While RGCs are operational in several countries, there is a lack of reported outcomes of this clinical model of care.5 11
The purpose of this study is to comprehensively determine the clinical utility and cost-effectiveness of genomic testing in patients with suspected GKD seen in a multidisciplinary RGC, and to compare this with their care and clinical diagnosis prior to referral. In addition, we aim to evaluate the value of a multidisciplinary RGC model from a patient perspective.
Study aims
To determine the proportion of patients with suspected GKD who obtain a positive diagnosis following genomic testing compared with their clinical diagnosis prior to testing.
To determine the clinical impact of genomic testing after 3 months’ follow-up in a cohort of patients with suspected GKD and identify subgroups of patients who are more likely to have a clinical impact following genomic testing.
To determine whether genomic testing in a cohort of patients with suspected GKD is cost-effective compared with usual care, and to identify whether genomic testing is cost-effective for specific subgroups.
To provide access to further research genomics participation for patients with suspected GKD who remain undiagnosed following clinical genomic testing.
To determine patient preferences regarding a service delivery model for a dedicated renal genetics service.
To determine the value of genomic testing in those with suspected GKD from a patient perspective.
Methods and analysis
Study design
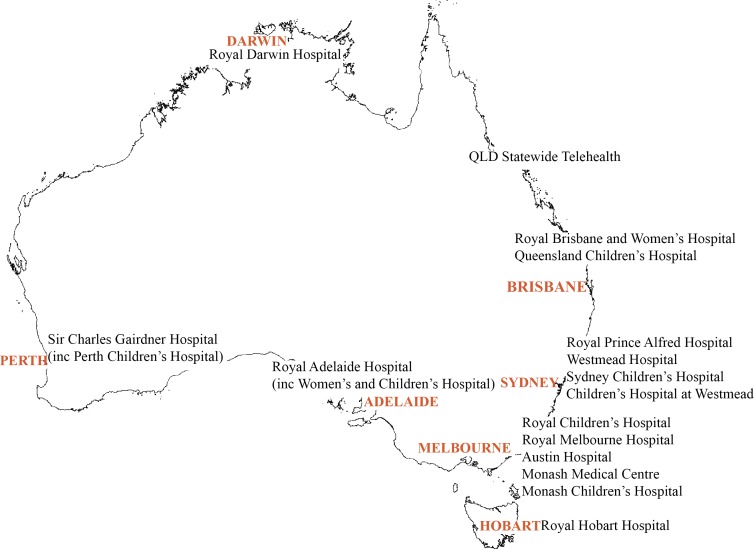
This prospective observational study will be undertaken at multiple Australian sites that provide multidisciplinary renal genetics services. There are 16 participating sites throughout Australia (figure 1). Patients who are referred by their treating physician to these multidisciplinary RGCs will be recruited over 4 years (figure 2). Patients may be referred for testing for a variety of reasons. These include confirmation of a suspected diagnosis, exclusion of differential diagnoses, clarification of mode of inheritance and for obtaining the diagnosis where one is previously unknown. For this reason, it was not possible to include a control arm, as this would prevent some patients from undergoing their usual clinical care. In order to enable determination of comparative clinical utility, planned diagnostic investigations will be nominated by the recruiting clinicians. In addition, data regarding any changes in management will be collected from the referring nephrologist at 3 months following the return of genomic testing results.
Figure 1.
Participating renal genetics clinics.
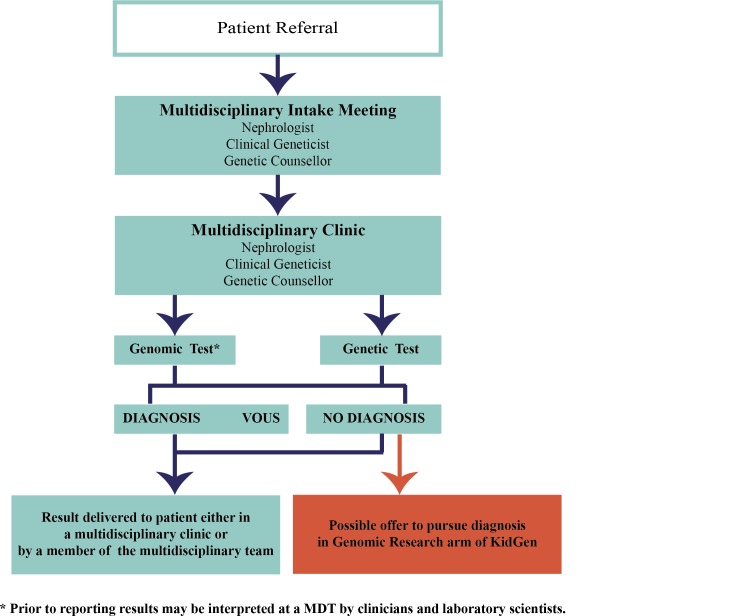
Figure 2.
Patient flow within a KidGen Renal Genetics Clinic. MDT, multidisciplinary team.
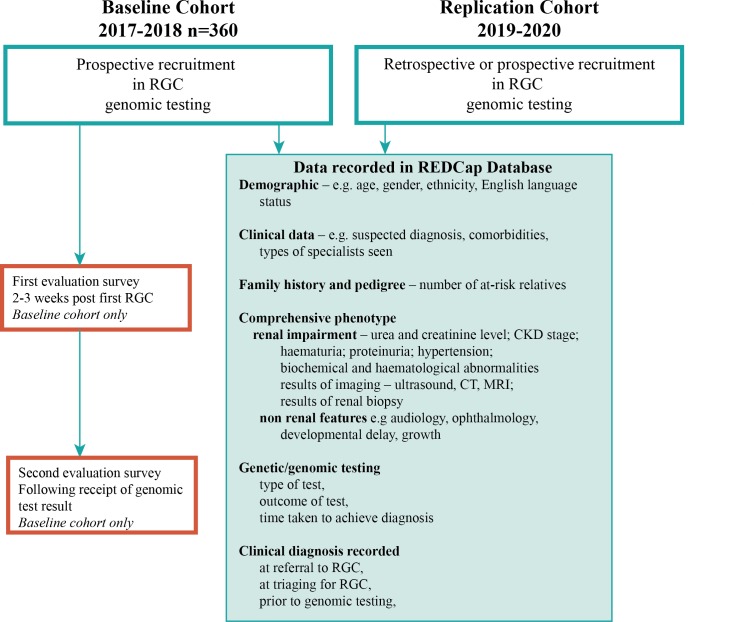
Recruitment of a baseline cohort of 360 participants occurred between 2017 and 2018 (figure 3) and a further mutually exclusive replication cohort will be recruited between 2019 and 2020. The replication cohort will have the same data collected but will also include patients with a prior genomic diagnosis. These patients will not participate in evaluation surveys (described below) but will be evaluated against all other outcome measures in order to further identify patient, disease and subcohort clinical outcomes enabled by larger scale and longer time span observation. The cohort of patients is anticipated to be ethnically diverse with a broad spectrum of renal phenotypes, clinical diagnoses and severity of kidney disease. Patients seen in a multidisciplinary RGC as part of standard clinical care will be invited to participate in the study if there is consensus of opinion by the clinic team that their kidney disease is likely genetic in origin. Patients without a clear clinical indication for prospective clinical genomic testing will be excluded in the baseline cohort, including an existing molecularly confirmed genetic diagnosis and those with heterogeneous or complex diseases with a low or unlikely monogenic diagnosis rate by current genomic sequencing (such as isolated congenital abnormalities of the kidney and urinary tract).12 Patients who undergo only Sanger sequencing are excluded from the study as this would not represent a genomic approach to clinical investigation and may instead represent clarification or segregation of an already identified familial variant. Patients who attend an RGC but do not undergo genetic testing through the clinic are also excluded from the study. This includes patients who decline clinically indicated testing. Written informed consent, both for clinical genomic testing and participation in the research study, will be obtained by the nominated clinical geneticist, nephrologist or genetic counsellor at each site.
Figure 3.
Recruitment workflow within the KidGen Renal Genetics Clinic network. CKD, chronic kidney disease; REDCap, Research Electronic Data Capture; RGC, renal genetics clinics.
Participant identifiers
All participants will be assigned a unique identifier for the purposes of this study at recruitment. Study data will be collected and managed using REDCap13 electronic data capture tools hosted at Murdoch Children’s Research Institute. Access to patient identifying details is restricted to clinicians providing care and to those who need contact details for evaluation purposes.
Measures
Once consented to the study, baseline demographic information, clinical information and detailed phenotypic information will be recorded by clinicians in REDCap. Demographic data including age, gender, ethnicity and English language status will be captured for all recruited patients. Further demographic information will be captured in patient surveys. Clinical data include suspected clinical diagnosis, comorbidities and types of specialists previously seen. A detailed family history and pedigree will be collected to identify the number of probands and at-risk relatives presenting for assessment. Comprehensive phenotypic information will also be collected including the presence of extrarenal features, renal impairment, urea and creatinine level, CKD stage, presence and details of hypertension, haematuria, proteinuria and biochemical/haematological abnormalities, results of imaging including ultrasound, CT and MRI, and results of renal biopsy if already performed. Data will also be collected regarding type of genomic test, outcomes of test and time taken to achieve diagnosis. Clinical diagnosis will be recorded as listed in the referral of each patient to the RGC, at the time of triaging for the RGC, prior to genomic testing and at the time of return of genomic testing results. This will enable comparison across multiple time points potentially leading up to a molecular genetic diagnosis.
Patient and public involvement
The project was reviewed, edited and revised after in-depth engagement with the Australian Genomic Health Alliance Consumer and Community Advisory group. Previous informal engagement with relevant representative local Australian kidney health patient organisations, including Polycystic Kidney Disease Australia and Kidney Health Australia, also occurred. Results from this study will be published and be accessible to all patients on request.
Evaluation surveys
Evaluation surveys will be distributed to be completed either online or on paper. Adult and proxy versions are available (parent for child). Participants are asked to complete the first survey at 2–3 weeks after attending the RGC at which they are offered and consent to testing, and the second following their receipt of genomic test results. The surveys were developed based on existing surveys in use in the Melbourne Genomics Health Alliance.
The first survey (table 1) captures additional demographic information and also includes questions about the experience of the consent process and views on the benefits of the multidisciplinary RGC. The Genetic Counselling Outcomes Survey14 is included in surveys 1 and 2 to evaluate genetic counselling outcomes of the multidisciplinary clinic. Knowledge questions and a question on the likelihood of the test finding the cause of the kidney disease are included in survey 1 to gauge understanding. The first survey includes closed and open questions exploring hopes and expectations for testing. Willingness-to-pay questions designed specifically for this study are included in both surveys to gauge value. Validated multiattribute quality of life instruments (12-Item Short Form Health Survey (SF-12) for adults and parents,15 Child Health Utility 9D instrument for children16) are included in surveys 1 and 2 to assess health-related quality of life. The Pediatric Quality of Life Inventory17 family impact module is included in surveys 1 and 2 for parents to assess family impact. Both surveys include questions on family planning.
Table 1.
Summary of survey measures
| Measure | Description | S1 | S2 |
| Demographics | Age, gender, marital status, education, income, number of dependents in household, postcode, private health insurance status. | X | |
| Patient-reported outcome measures | CHU9D and PedsQL family impact for paediatric surveys OR SF-12 for adult surveys |
X | X |
| Experience of the multidisciplinary clinic | Three study-specific questions exploring advantages and disadvantages of multidisciplinary renal genetics clinics. | X | X |
| Genetic counselling | 24-item scale measuring outcomes of genetic counselling GCOS-24. | X | X |
| Family planning | Four study-specific questions addressing plans for another child, estimated recurrence of the kidney condition, concern about recurrence, interest in reproductive technologies (parent surveys only). | X | |
| Understanding | In survey 1, eight study-specific questions address participant understanding of: types of potential results (four questions), potential familial implications (one question), ways in which the data can be used (two questions) and number of genes examined (one question). In survey 2, two study-specific questions used to measure recall and understanding of result. |
X | X |
| Willingness to pay (value) | Study-specific questions included to establish a quantitative reference for the value placed on testing. | X | X |
| Information provision | Study-specific questions to assess participant perception about the way in which information (3 items—S1) and results (3 items—S2) were provided. | X | X |
| Hopes/expectation | Eight study-specific questions exploring participants’ reasons for agreeing to the test, rated on a 5-point scale as extremely unimportant to extremely important. | X | |
| Likelihood | One study-specific question to determine participant’s perception of the likelihood testing will find the cause of the condition. | X | |
| Decision regret | 5-item scale measuring distress or remorse after a (healthcare) decision (ref O’Connor).18 | X | |
| Value of the test | Eleven study-specific questions exploring the value to participants of having had the test rated on a 4-point scale as not valuable to extremely valuable or not applicable. | X | |
| Impact of the test | Two study-specific questions asking about the impact of the test on family planning. | X |
CHU9D, Child Health Utility 9D instrument; GCOS-24, Genetic Counselling Outcome Scale; PedsQL, Pediatric Quality of Life Inventory; SF-12, 12-Item Short Form Health Survey.
In addition to the measures described above, the second survey includes study-specific questions assessing understanding of the genomic results, the impact this will have on the patient and their family and perceived value of genomic testing. The Decision Regret Scale18 is also included in survey 2.
Sequencing
Genomic sequencing will be undertaken and reported in clinically accredited laboratories. It is envisaged that these are likely to include but not be limited to the National Association of Testing Authorities accredited diagnostic laboratories at Children’s Hospital at Westmead, Victorian Clinical Genetics Service, Genome. One, SA Pathology and PathWest. The specific clinical test requested for each participant will be selected by the treating clinician/s at the attending RGC according to clinical indication. The spectrum of genomic testing that will be employed is anticipated to include targeted exome, whole exome and whole genome sequencing, including the application of virtual gene panels based on patient phenotype. When additional copy number variation assessment and/or variant confirmation is indicated, then multiplex ligation-dependent probe application, chromosomal microarray or Sanger sequencing may be undertaken in addition to genomic sequencing. The cost per test is expected to be $A1200–$A2400 depending on the specific test and diagnostic provider, with individual test costs to be ascertained directly. Further, each participant will have genomic sequencing performed once unless technical or sequence quality issues require resequencing to enable clinical reporting. Based on clinical indication, further analysis of disease or phenotype-specific virtual gene panels may occur, with any such sequence reanalysis being recorded.
All participants will provide written clinical consent for genomic testing and analysis will be restricted to the assessment of genes related to the condition of interest. In this instance, analysis will be limited to a predefined gene list of 360 genes related to nephropathy.19 Secondary findings unrelated to the presenting condition will not be reported in this study. Written informed consent for participation in the research study will be attained separately, which includes options to consent to accessing the Medicare Benefits Schedule (MBS) and Pharmaceutical Benefits Scheme (PBS), hospital and emergency data sets and data sharing in addition to study evaluation data collection. The MBS and PBS are listings of all medical services and medicines subsidised by the Australian government, respectively. In addition, an optional written consent will be obtained in order to share data and samples for use in ethically approved research outside of the study.
Health economic evaluation
An economic evaluation will be conducted to assess the incremental cost-effectiveness of NGS compared with usual care in patients with suspected kidney disease. A microsimulation model of disease progression will be developed to estimate the lifetime costs and outcomes associated with usual care and NGS. The analysis will be conducted from an Australian healthcare system perspective in line with recommended practices,20–22 and based on the outcomes of cost per additional diagnosis, cost per quality-adjusted life year and net monetary benefit.
Hospital patient-level resource use and cost data will be acquired for each study participant to cost the diagnostic and short-term medical management in the NGS arm. The additional per-patient diagnostic investigations that could potentially be incurred in the usual care pathway will be identified based on a review of national and international guidelines on the diagnosis of suspected kidney disease and clinical expertise from each of the 16 participating centres across Australia. The Australia and New Zealand Dialysis and Transplant Registry will be used to undertake a survival analysis of patients with CKD. Parametric and non-parametric methods will be used to estimate transition probabilities for key endpoints, including: all-cause mortality, initiation of dialysis and transplantation. This will allow for the development of evidence-based transition probabilities while controlling for patient-specific factors such as age, gender, treatment pathways and disease status. Unit costs associated with these endpoints and related treatments will be drawn from established national sources.23–25 A microsimulation will be used to capture the costs and benefits associated with earlier renal replacement therapy and delayed dialysis from the introduction of NGS. To accurately model the implications, registry-derived transition probabilities will be adjusted, on the basis of published evidence,26 to reflect the impact of new treatment pathways following NGS.
For the cost-utility analysis, participants’ responses to the SF-12 measure will initially be used to generate utilities at baseline prior to NGS and following the return of NGS findings. A review of the literature will be undertaken to complement these estimates with evidence from secondary sources, such as utilities for CKD27 and renal transplantation.28 The cost-benefit analysis will rely on a contingent valuation exercise that was undertaken to understand the personal value of NGS to the families. Deterministic and probabilistic sensitivity analyses will be undertaken to explore the robustness of the findings to plausible variations in key assumptions around costs, outcomes and transition probabilities, and to consider the broader issue of generalisability of the study’s results. Cost-effectiveness acceptability curves will be generated to reflect decision uncertainty.
Study outcomes
The primary outcome will be the proportion of patients who receive a molecular diagnosis via genomic testing (diagnostic rate). Secondary outcomes will be change in clinical diagnosis following genomic testing, change in clinical management following genomic testing and cost-effectiveness of genomic testing compared with non-genomic diagnostic investigations. Survey outcomes will include the proportion of patients who preferred to be seen by the multidisciplinary team compared with those who preferred to be seen in individual clinics. In addition, the Genetic Counselling Outcome Scale will be compared before and after testing and between those with a positive/negative genomic testing result.
Data analysis
Results on diagnostic utility and clinical implications will be expressed as frequencies and percentages for categorical variables and be compared using the χ2 test. Continuous non-normally distributed variables will be expressed as median (IQR) and compared using the Mann-Whitney test. Normally distributed continuous variables will be expressed as mean±SD and compared using the Student’s t-test. Multivariable logistic regression will be performed to determine which variables may predict a positive genomic diagnosis, such as age of onset of CKD, aetiology of CKD, family history or presence of haematuria/proteinuria/cysts. This will help develop professional guidelines regarding genomic testing in suspected GKD. In addition, the diagnostic rates between different genomic testing modalities will also be compared. The target sample size is 500 patients across both baseline and replication cohorts.
Discussion
In this protocol, we describe the rationale and methods for a prospective observational study of a national cohort of patients with suspected genetic renal disease that are referred to multidisciplinary RGCs throughout Australia. The strengths of this study are the multicentre design, prospective data collection and a real-world clinical setting. The absence of a control arm is a limitation of this study, however it was not feasible to randomise patients in this study for two reasons. First, genomic testing is clinically indicated and performed routinely in most participating states/centres, and it would therefore be unethical to deny genomic testing to some patients where it is clinically indicated and current standard of care. Second, feedback from patient representatives, the community and genetic counsellors highlighted that it was unacceptable to deny testing to some patients for the purpose of a control arm. Further, those patients not receiving a genomic test after clinical assessment are likely to do so in a non-random fashion thus precluding their analysis as a true control group.
Genomic technologies have transformed the concept of precision medicine in many specialties, however the potential benefits in kidney medicine are yet to be demonstrated. Until now, the diagnostic utility of genomic testing has been assessed in a small number of patients or in a research context.29 30 In addition, while studies which perform genomic sequencing in larger cohorts6 31 are emerging, detailed phenotypic information is not being collected and there remains a paucity of data on clinical outcomes. Importantly, these large-scale studies include patients with all types of CKD and are thus powered to understand prevalence of GKD rather than to determine utility and yield in a cohort in which the clinician suspects genetic disease.6 Data on genomic sequencing in a real-world clinical environment at a national scale for a targeted population of patients with suspected GKD are lacking and the health economic impacts of genomic testing are not well understood or established.32 We believe that collecting such detailed information from a prospectively ascertained cohort will enable us to determine the diagnostic yield and comprehensive clinical implications, to inform future practice recommendations.
To our knowledge, this is the first time that a structured, objective and minimally bias approach is undertaken to measure the outcomes of genomic testing in kidney disease. Results from this study will provide the opportunity to determine the clinical implications of genomic testing in a large cohort of patients, and further enable analysis of which subgroups of patients may benefit most. Furthermore, due to the pragmatic nature of this study, these results more likely to be replicated in a clinical environment. By assessing the utility, cost-effectiveness and implementation aspects of genomic testing in patients with kidney disease, the results of this study will inform patients, treating physicians and health services and define priorities for future trials. Collectively, this is anticipated to have significant impacts on clinical practice and health service redesign.
Ethics and dissemination
Ethics
Ethics approval for this project has been obtained by Australian Genomics Health Alliance as part of the research study, Australian Genomics Health Alliance: Preparing Australia for Genomic Medicine, and issued by Melbourne Health HREC/16/MH/251. Governance site-specific approval for the project has been obtained for each of the participating clinic sites. All participants will provide written informed consent for data collection and to undergo clinically relevant genetic/genomic testing.
Dissemination plan
The main findings of this study will be published in peer-reviewed journals and will also be presented at national and international conferences. We will also issue reports of results of the study to the Australian Government, State/Territory Governments and health organisations in order to inform future policy and guidelines. This study will also contribute to the training and development of postdoctoral students.
Supplementary Material
Footnotes
Contributors: KJ contributed to the design of the study and drafted the manuscript. CQ and AJM conceived the project and obtained funding for the clinical flagships and research study, made substantial contributions to the design of the study and were major contributors in drafting the manuscript. LW and ZS undertook significant project design and data management elements, provided specific design elements around outcome analysis and were major contributors in drafting the manuscript. IG, YW and MM provided specific design elements around health economic analysis. SB provided specific implementation design elements. AM, HM, RF, AC, MS, MJ, PK and CP contributed to regionalised study design and implementation. All authors contributed to the drafting of the manuscript, read and approved the final manuscript. In addition to the authors listed, the broader KidGen Collaborative scientific, clinical and diagnostic membership had input into this protocol.
Funding: Operating as the KidGen Renal Genetics Flagship, this work was supported by the Melbourne Genomics Health Alliance (Melbourne Genomics) and grants from the Royal Children’s Hospital Foundation and by the NHMRC Genomics TCR Grant 1113531, entitled Preparing Australia for Genomic Medicine: A proposal by the Australian Genomics Health Alliance. Melbourne Genomics is funded by the 10 members and the State Government of Victoria (Department of Health and Humans Services). In-kind funding support was provided for renal genetics clinic operation and genomic test provision by Queensland Health (Metro North Hospital and Health Service; Children’s Health Queensland Hospital and Health Service), New South Wales Health, South Australia Health, Western Australia Department of Health, Northern Territory Department of Health and Tasmanian Department of Health and Human Services.
Map disclaimer: The depiction of boundaries on the map(s) in this article do not imply the expression of any opinion whatsoever on the part of BMJ (or any member of its group) concerning the legal status of any country, territory, jurisdiction or area or of its authorities. The map(s) are provided without any warranty of any kind, either express or implied.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: Ethics approval for this project has been obtained by Australian Genomics Health Alliance as part of the research study, Australian Genomics Health Alliance: Preparing Australia for Genomic Medicine, and issued by Melbourne Health HREC/16/MH/251. Governance site-specific approval for the project has been obtained for each of the participating clinic sites.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Mallett A, Patel C, Salisbury A, et al. The prevalence and epidemiology of genetic renal disease amongst adults with chronic kidney disease in Australia. Orphanet J Rare Dis 2014;9:98 10.1186/1750-1172-9-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vivante A, Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol 2016;12:133–46. 10.1038/nrneph.2015.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rabbani B, Mahdieh N, Hosomichi K, et al. Next-Generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet 2012;57:621–32. 10.1038/jhg.2012.91 [DOI] [PubMed] [Google Scholar]
- 4. Groopman EE, Rasouly HM, Gharavi AG. Genomic medicine for kidney disease. Nat Rev Nephrol 2018;14:83–104. 10.1038/nrneph.2017.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mallett A, Fowles LF, McGaughran J, et al. A multidisciplinary renal genetics clinic improves patient diagnosis. Med J Aust 2016;204:58–9. 10.5694/mja15.01157 [DOI] [PubMed] [Google Scholar]
- 6. Groopman EE, Marasa M, Cameron-Christie S, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 2019;380:142-151 10.1056/NEJMoa1806891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bresin E, Daina E, Noris M, et al. Outcome of renal transplantation in patients with non-Shiga toxin-associated hemolytic uremic syndrome: prognostic significance of genetic background. Clin J Am Soc Nephrol 2006;1:88–99. 10.2215/CJN.00050505 [DOI] [PubMed] [Google Scholar]
- 8. Sellier-Leclerc A-L, Fremeaux-Bacchi V, Dragon-Durey M-A, et al. Differential impact of complement mutations on clinical characteristics in atypical hemolytic uremic syndrome. J Am Soc Nephrol 2007;18:2392–400. 10.1681/ASN.2006080811 [DOI] [PubMed] [Google Scholar]
- 9. Gross O, Licht C, Anders HJ, et al. Early angiotensin-converting enzyme inhibition in Alport syndrome delays renal failure and improves life expectancy. Kidney Int 2012;81:494–501. 10.1038/ki.2011.407 [DOI] [PubMed] [Google Scholar]
- 10. Varner JD, Chryst-Stangl M, Esezobor CI, et al. Genetic testing for Steroid-Resistant-Nephrotic syndrome in an outbred population. Front Pediatr 2018;6 10.3389/fped.2018.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alkanderi S, Yates LM, Johnson SA, et al. Lessons learned from a multidisciplinary renal genetics clinic. QJM 2017;110:453–7. 10.1093/qjmed/hcx030 [DOI] [PubMed] [Google Scholar]
- 12. Nicolaou N, Pulit SL, Nijman IJ, et al. Prioritization and burden analysis of rare variants in 208 candidate genes suggest they do not play a major role in CAKUT. Kidney Int 2016;89:476–86. 10.1038/ki.2015.319 [DOI] [PubMed] [Google Scholar]
- 13. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McAllister M, Wood AM, Dunn G, et al. The genetic counseling outcome scale: a new patient-reported outcome measure for clinical genetics services. Clin Genet 2011;79:413–24. 10.1111/j.1399-0004.2011.01636.x [DOI] [PubMed] [Google Scholar]
- 15. Brazier JE, Roberts J. The estimation of a preference-based measure of health from the SF-12. Med Care 2004;42:851–9. 10.1097/01.mlr.0000135827.18610.0d [DOI] [PubMed] [Google Scholar]
- 16. Furber G, Segal L. The validity of the child health utility instrument (CHU9D) as a routine outcome measure for use in child and adolescent mental health services. Health Qual Life Outcomes 2015;13:22 10.1186/s12955-015-0218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glascoe FP. Using parents' concerns to detect and address developmental and behavioral problems. J Soc Pediatr Nurs 1999;4:24–35. 10.1111/j.1744-6155.1999.tb00077.x [DOI] [PubMed] [Google Scholar]
- 18. Brehaut JC, O'Connor AM, Wood TJ, et al. Validation of a decision regret scale. Med Decis Making 2003;23:281–92. 10.1177/0272989X03256005 [DOI] [PubMed] [Google Scholar]
- 19. Little MH, Quinlan C. Advances in our understanding of genetic kidney disease using kidney organoids. Pediatr Nephrol 2019. 10.1007/s00467-019-04259-x. [Epub ahead of print: 07 May 2019]. [DOI] [PubMed] [Google Scholar]
- 20. Husereau D, Drummond M, Petrou S, et al. Consolidated Health Economic Evaluation Reporting Standards (CHEERS)--explanation and elaboration: a report of the ISPOR Health Economic Evaluation Publication Guidelines Good Reporting Practices Task Force. Value Health 2013;16:231–50. 10.1016/j.jval.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 21. Philips Z, Ginnelly L, Sculpher M, et al. Review of guidelines for good practice in decision-analytic modelling in health technology assessment 2004:iii-iv–ix p. [DOI] [PubMed]
- 22. Committee PBA Guidelines for preparing submissions to the Pharmaceutical Benefits Advisory Committee : Health AGDo. 5.0 ed Canberra: Australian Government Department of Health, 2016. [Google Scholar]
- 23. Australia M Medicare Australia. pharmaceutical benefits schedule item reports. Health Insurance Commission, 2018. [Google Scholar]
- 24. Australia M Medicare benefits schedule (Mbs) item statistics reports. Health Insurance Commission, 2018. [Google Scholar]
- 25. Government A National Hospital cost data collection cost report round 20 (2015-2016). Report. Canberra, Australia, 2015-2016. [Google Scholar]
- 26. ANZDATA The 40th annual report: ANZDATA Registry report. Adelaide: Australia and New Zealand Dialysis and Transplant Registry, 2017. [Google Scholar]
- 27. Gorodetskaya I, Zenios S, McCulloch CE, et al. Health-Related quality of life and estimates of utility in chronic kidney disease. Kidney Int 2005;68:2801–8. 10.1111/j.1523-1755.2005.00752.x [DOI] [PubMed] [Google Scholar]
- 28. Laupacis A, Keown P, Pus N, et al. A study of the quality of life and cost-utility of renal transplantation. Kidney Int 1996;50:235–42. 10.1038/ki.1996.307 [DOI] [PubMed] [Google Scholar]
- 29. Sadowski CE, Lovric S, Ashraf S, et al. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 2015;26:1279–89. 10.1681/ASN.2014050489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heidet L, Morinière V, Henry C, et al. Targeted Exome Sequencing Identifies PBX1 as Involved in Monogenic Congenital Anomalies of the Kidney and Urinary Tract. J Am Soc Nephrol 2017;28:2901–14. 10.1681/ASN.2017010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lata S, Marasa M, Li Y, et al. Whole-Exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med 2018;168:100–9. 10.7326/M17-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med 2018;20:1122–30. 10.1038/gim.2017.247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.