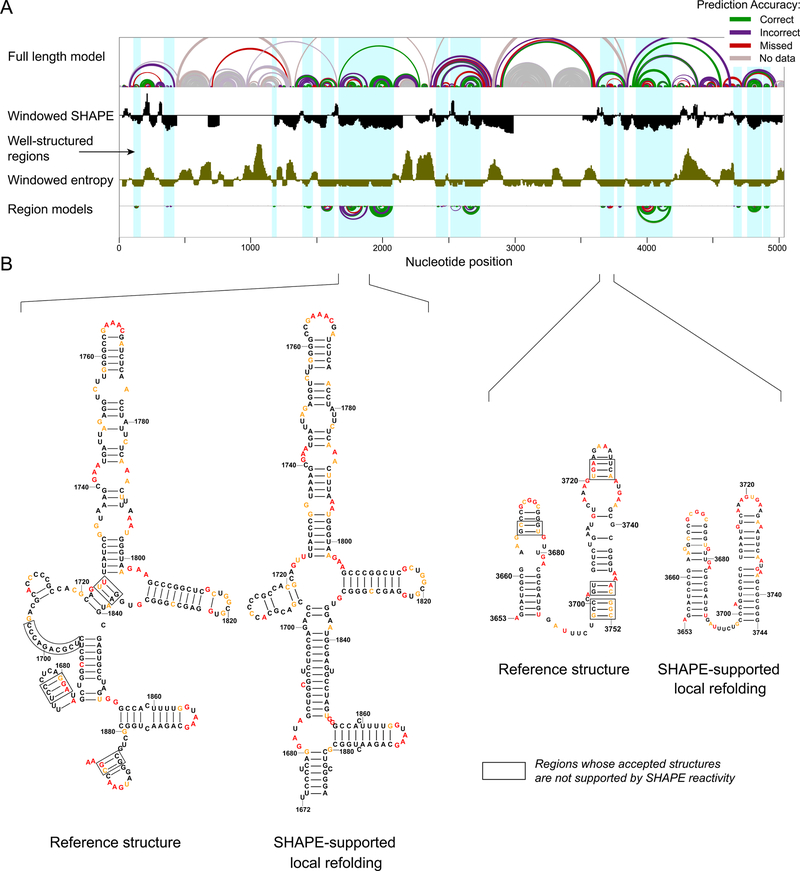
Figure 4. Structural characterization of the human 28S rRNA.
(A) (top) Model of the full-length 28S rRNA with arcs representing correct, incorrect, and missing base pairs relative to the accepted structure. (middle) SHAPE reactivity (black) and entropy (brown) shown as medians over 51-nt centered windows along the full-length 18S rRNA with the axes crossing at 0.3 and 0.08, respectively. (bottom) SHAPE-directed structure models for lowSS regions modeled as independent elements. (B) Nucleotide-level models of selected regions, highlighting plausible SHAPE-supported local refolding. Local structures that are clearly not compatible with the accepted structure are boxed (defined as nucleotides with high reactivity in base paired regions or low reactivity in consecutive unpaired regions). Nucleotides are colored by SHAPE reactivity (legend in Fig. 1).