Fig 2. Host and MHV68 miRNAs bind frequently to the CDS or 3’ UTR of target transcripts, but MHV68 miRNAs cumulatively display a non-canonical binding profile.
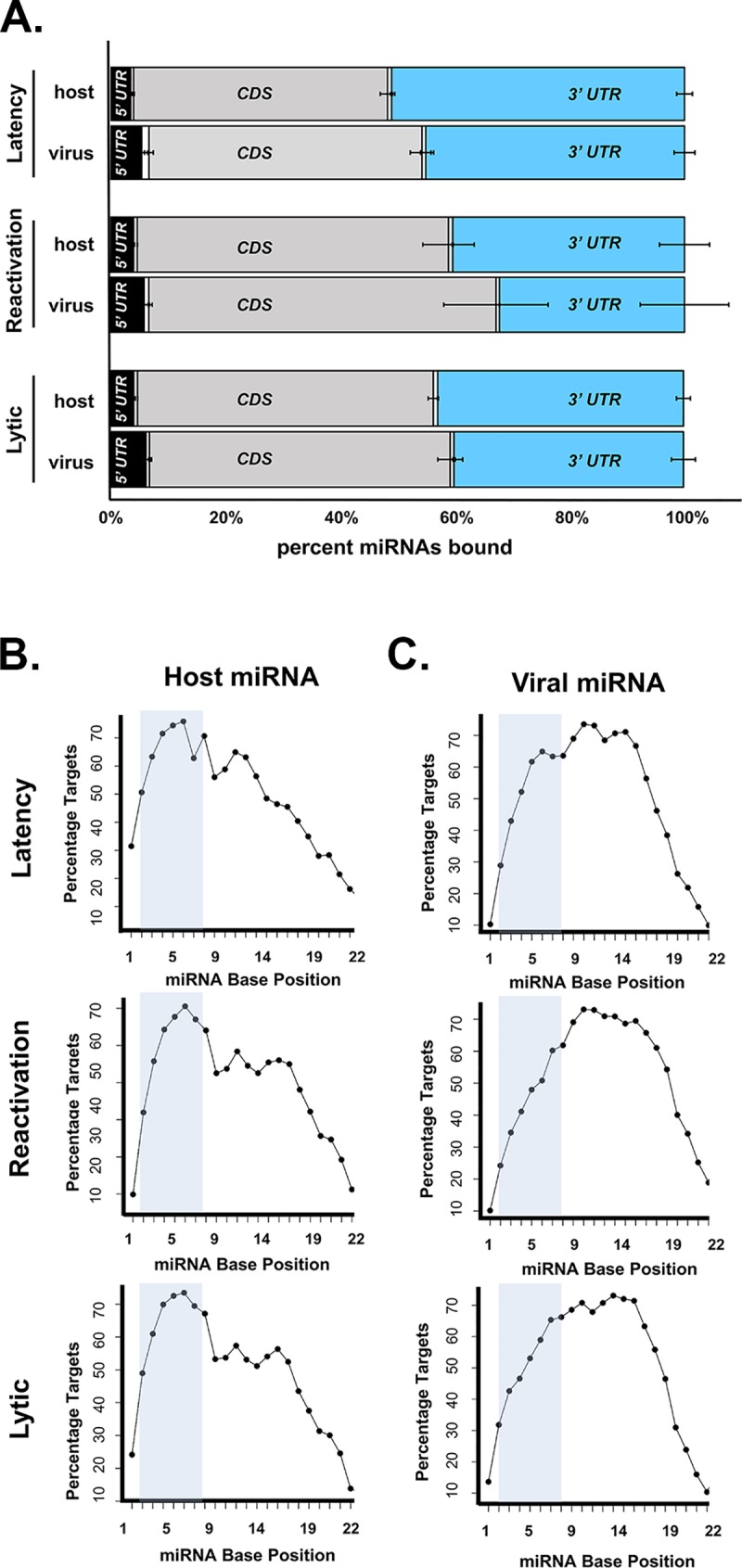
A. For all miRNA-mRNA hybrids within each infection condition, the percentage of total host or viral miRNAs bound to each region of target transcripts is indicated. Regions were categorized as 5’ UTR, 5’ UTR-CDS, CDS, CDS-3’ UTR, or 3’ UTR. Error bars represent the standard deviation across three biological replicates. The target transcript 3’ UTR (the canonical miRNA binding region) is highlighted in blue. B, C. For all miRNA hybrids within each infection condition, the percentage of target mRNAs that bound to individual nucleotides along the length of each miRNA was determined. Percentages were calculated for all host (B) or virus (C) miRNA-mRNA interactions within cumulative latency, reactivation and lytic datasets.
