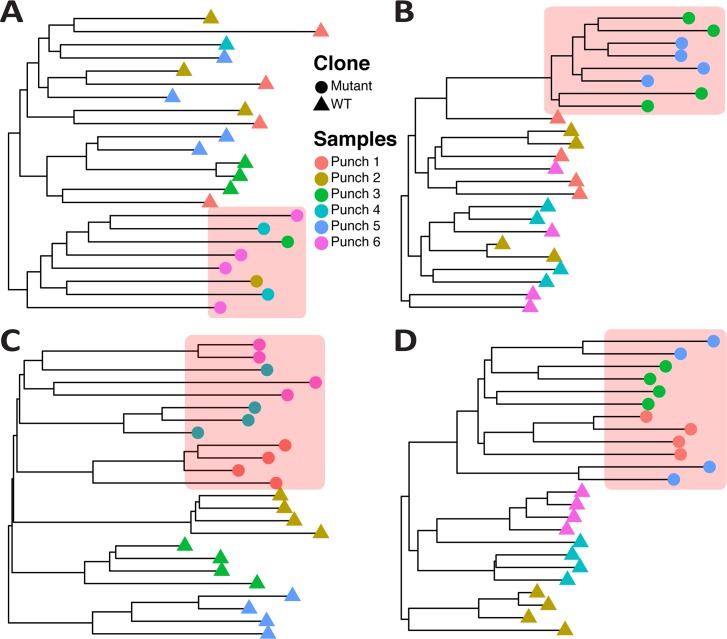
Fig 5. Biases of single-cell sequencing when cells are taken from spatially separated bulk samples.
Whereas taking N random cells from a tumour highly reduces sampling bias, this is often not how single-cell from neoplasms are sampled. Often first small chunks of the tumour are dissected and then single-cells are isolated from those. (A) neutral homogeneous, (B) selective homogeneous, (C) neutral boundary driven, (D) selective boundary driven. For each of our representative examples, we simulated this type of sampling and show how this impacts severely on the phylogenetic tree and patterns of clonal intermixing. In particular, single-cell sampling from bulks alters the detected phylogenetic relationship of the cells because, since groups of cells come from spatially segregated regions, those appear more closely related than expected by chance. This is an important source of sampling bias that needs to be considered when analysing single-cell phylogenies. Cells coming from the ‘red’ mutant subclone are highlighted in the red shaded box.