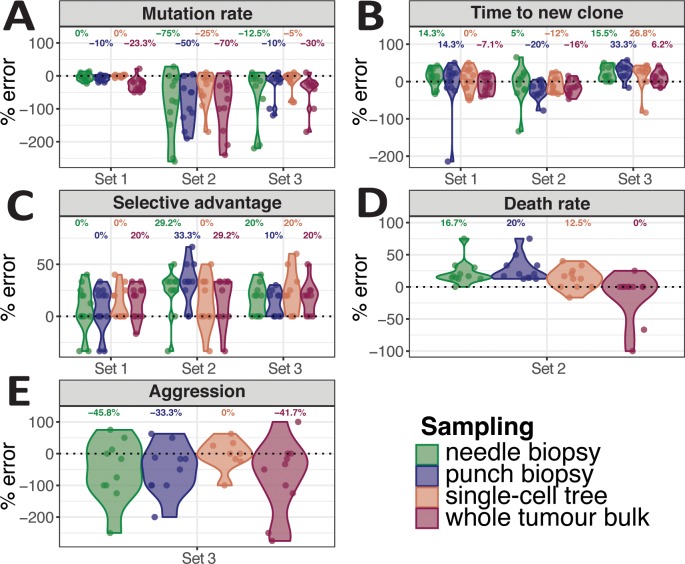
Fig 6. Statistical inference framework to recover evolutionary parameters.
We combined our model with a statistical inference framework (Approximate Bayesian Computation–Sequential Monte Carlo) in order to infer the evolutionary parameters of selection and growth from the data. We tested this framework on 34 synthetic (target) tumours for which we generated genomic data. Our of these 34 target cases, 13 were characterised by homogeneous growth with no cell death (A, Set 1), 11 were homogeneous but with cell death (B, Set 2), and 10 where characterised by boundary driven growth (C, Set 3), see all parameters in S1 Table. We tested the ability to recovery parameters of 4 different sampling schemes: punch samples, needle biopsies, single cell phylogenetic trees and whole-tumour sampling (see Materials and Methods for details). We report the percentage error of the inference (true parameter value–inferred value based on the mode of the posterior probability) for each parameter and scenario. See prior parameter ranges in S2 Table. (D) For the homogeneous stochastic cell death scenario (Set 2), we also report the error in recovering the death rate parameter d. (E) For the boundary driven growth scenario we report the error in recovering boundary driven growth parameter a (Set 3).