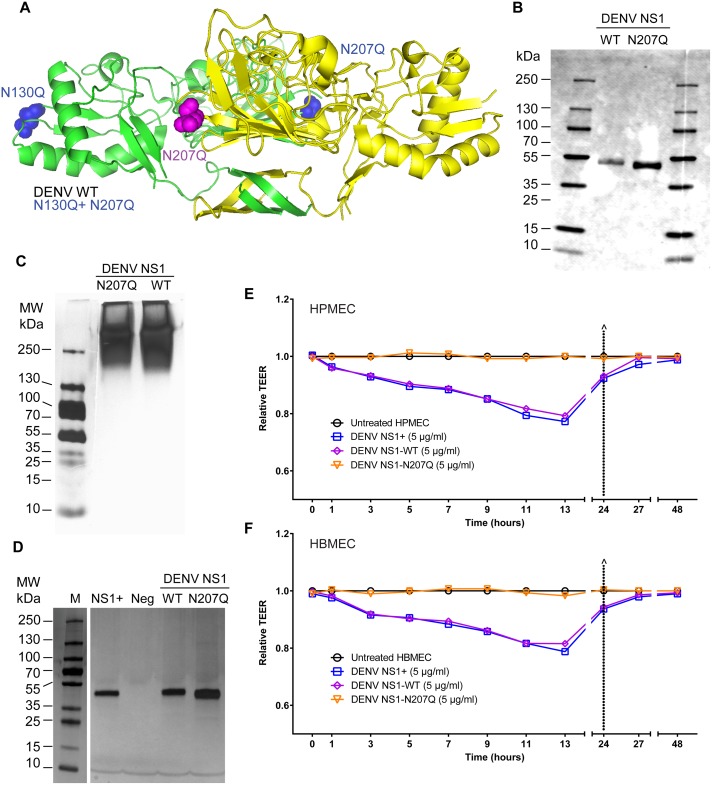
Fig 1. Mutation of the N-glycosylation site 207 prevents DENV NS1-induced hyperpermeability of endothelial cells.
(A) Structural model of a DENV2 NS1 dimer showing site-specific mutations. Color-coding highlights two NS1 monomers (one in green, one in yellow) and amino acid changes in corresponding constructs (blue highlights changes in the green monomer; pink highlights changes in the yellow monomer). WT, N130Q, N207Q, and N130Q+N207Q are all on the DENV2 NS1 backbone. PDB ID: 4O6B [14]. (B) Western blot of SDS-PAGE of in-house purified and dialyzed DENV NS1-WT and NS1-N207Q mutant, showing monomeric form, using anti-NS1 mAb (7E11) for detection. (C) Silver stain of native PAGE showing oligomeric forms of both purified and dialyzed in-house DENV NS1-WT and NS1-N207Q mutant (MW: >250 kDa). (D) Silver stain of SDS-PAGE gels of purified and dialyzed in-house DENV NS1-WT and NS1-N207Q NS1 mutant, showing purity, using the Pierce Silver Stain Kit. Positive control, NS1+, DENV NS1 from Native Antigen Company; Neg, dialyzed elution buffer. Molecular weight marker, 10-250-kDa protein ladder. (E-F) DENV NS1 triggers endothelial permeability of HPMEC (E) and HBMEC (F) monolayers evaluated by TEER (Ohms) at the indicated time-points. Relative TEER was calculated as previously described [8]. Data are representative of 2–3 individual experiments. NS1+, NS1 from Native Antigen Company.