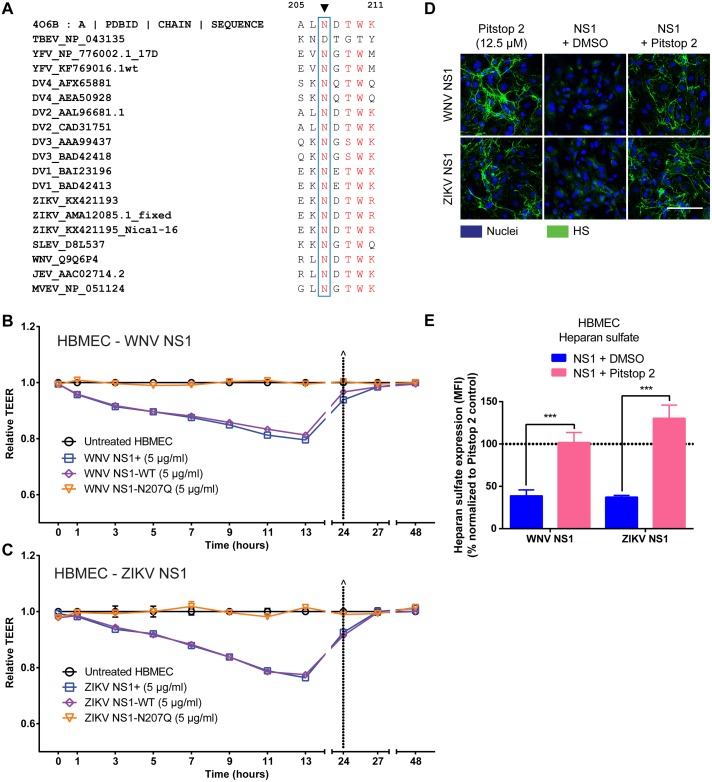
Fig 8. Mutation of the N207 glycosylation site abrogates endothelial hyperpermeability induced by NS1 proteins from WNV and ZIKV.
(A) Amino acid sequence alignment of flavivirus NS1 sequences indicate that N207 glycosylation site is highly conserved. Accession numbers for each NS1 protein sequences are listed after the corresponding flavivirus names. Sequence analysis was performed using MultAlin and recreated in Excel. The amino acid sequence of NS1 from aa 205–211 is shown. (B-C) Human brain microvascular endothelial cells (HBMEC) were grown on Transwell semi-permeable membranes (0.4 μm pore size), and WNV NS1 (B) or ZIKV NS1 (C) proteins (5 μg/ml) were added to the apical chamber (NS1+, commercially purchased NS1, blue squares; NS1-WT, produced in-house, purple diamonds; NS1-N207Q mutant, orange triangles). A TEER assay was used to evaluate the effect of these NS1 proteins on endothelial permeability at indicated time-points over 48 hours. (^) represents change of medium. Relative TEER values from 2 independent experiments performed in duplicate are plotted. Error bars indicate SEM. (D) The integrity of the EGL on HBMEC was assessed by the presence of heparan sulfate surface expression (green) at 6 hpt with WNV NS1 or ZIKV NS1 (5 μg/ml) in the presence or absence of Pitstop 2 (12.5 μM) at 37°C, as visualized via IFA. Nuclei were stained with Hoechst (blue). Images (20X; scale bars, 50 μm) are representative of 3 independent experiments. (E) Quantification of MFI in (D) from 3 independent experiments. ***, p < 0.001.