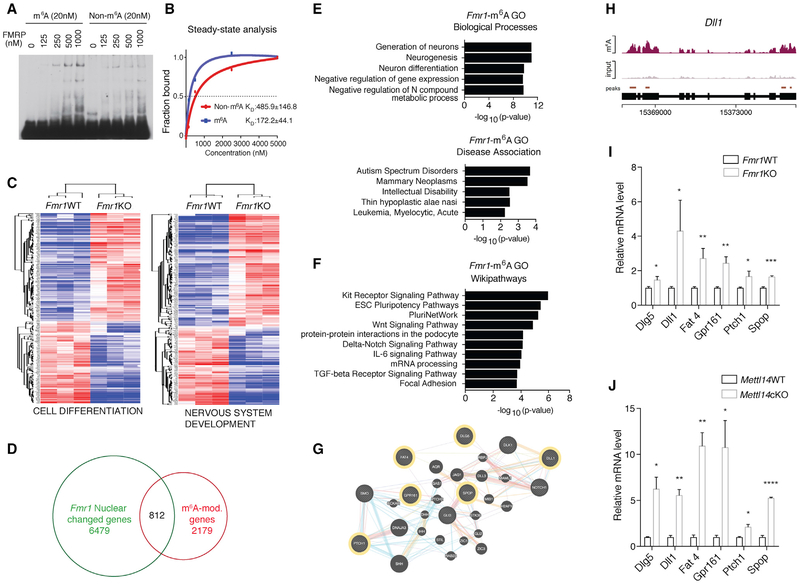
Figure 3. FMRP Preferentially Binds m6A-Modified RNAs to Promote Their Nuclear Export in Regulating Neural Differentiation.
(A) EMSA comparing FMRP binding to non-methylated (left) or methylated (right) RNA.
(B) Steady-state analysis of FMRP binding to methylated or non-methylated RNA using bio-layer interferometry. Results are averaged from three independent experiments.
(C) Heatmaps comparing WT and Fmr1KO NPC nuclear expression of genes related to cell differentiation (left) and nervous system development (right).
(D) Venn diagram showing RNAs that are m6A modified and differentially expressed in Fmr1KO nucleus.
(E) Gene Ontology (GO) analysis of RNAs that are both differentially expressed in Fmr1KO nucleus and m6A modified. Biological processes (top) and disease associations (bottom) are shown.
(F) Pathway analysis of RNAs that are both differentially expressed in Fmr1KO nucleus and m6A modified.
(G) Gene interaction network of Hedgehog- and Notch-related signaling components and their cross-talk. m6A-tagged FMRP targets are outlined in yellow.
(H) Coverage plot of m6A modification of Dll1 mRNA.
(I) RNAs of Hedgehog- and Notch-related components are retained in Fmr1KO nucleus quantified by qRT-PCR: Dlg5 (*p = 0.0324), Dll1 (*p = 0.0433), Fat4 (**p = 0.0065), Gpr161 (**p = 0.0020), Ptch1 (*p = 0.0262), and Spop (***p = 0.0006); n = 5 WT, 3 KO biological replicates. Normalization is to U1. Data are presented as mean + SEM.
(J) RNAs of Hedgehog- and Notch-related components are retained in Mettl14cKO nucleus quantified by qRT-PCR: Dlg5 (*p = 0.0180), Dll1 (**p = 0.0016), Fat4 (**p = 0.0021), Gpr161 (*p = 0.0361), Ptch1 (*p = 0.0164), and Spop (****p < 0.0001); n = 3 WT, 4 cKO biological replicates. Normalization is to U1. Data are presented as mean + SEM.