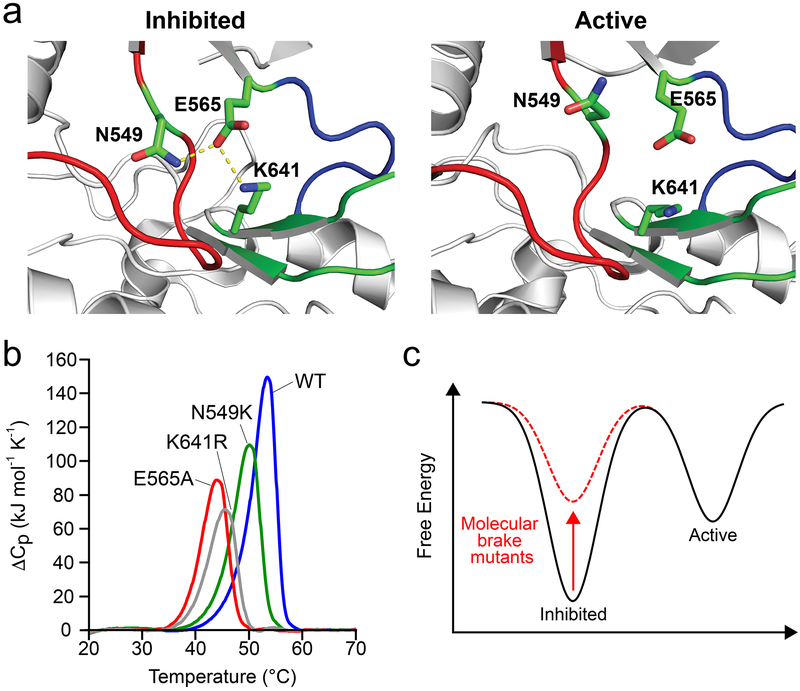
Figure 1. Gain-of-function mutations at the molecular brake destabilize the inhibited conformation.
(a) Left: structural view depicting engaged molecular brake as observed in the inhibited FGFR1 kinase (PDB: 3KY2); note that numbering follows that of FGFR2K. Right: disengaged molecular brake as seen in the A-loop tyrosine phosphorylated activated FGFR2K (PDB: 2PVF). Kinase molecule is shown as gray cartoon; αC-β4 loop, kinase hinge, and β8 strand contributing residues to the molecular brake are highlighted in red, blue and green, respectively. The triad of residues (i.e., N549, E565 and K641) comprising the molecular brake are shown as green sticks. Oxygen and nitrogen atoms are colored red and blue, respectively; hydrogen bonds are denoted by dashed yellow lines. (b) Differential scanning calorimetry thermogram data comparing thermal stabilities of wild-type FGFR2K and molecular brake mutants (i.e., N549K, E565A, and K641R). Note that molecular brake mutants have reduced melting temperatures relative to the wild-type FGFR2 kinase. A complete list of melting temperatures is shown in Table 1. (c) Schematic of free energy landscape in wild-type (black) and molecular brake mutants (dashed red). The red arrow highlights that molecular brake mutations elevate the basal free energy of the kinase; i.e., they destabilize the inhibited state.