Figure 2.
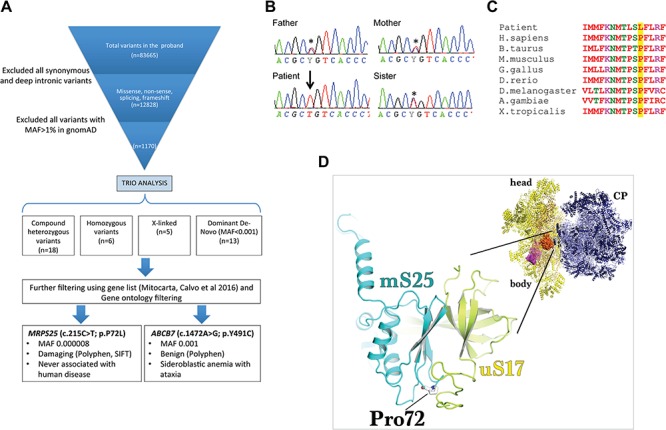
Identification of a potential pathogenic MRPS25 mutation by exome sequencing. (A) Filtering of the identified variants was performed using Varaft platform by comparing the proband’s and parents’ exomes. The analysis resulted in 42 genetic variants (de novo, recessive and X-linked), of which two were in genes encoding for mitochondrial proteins. Only the c.215C>T change in MRPS25 was predicting as damaging (using SIFT and Polyphen tools). (B) Sanger sequencing confirmed that each parent and the healthy sister were heterozygous (asterisk) for the mutation in MRPS25 while the affected patient was homozygous (arrow). (C) Sequencing alignment of the human MRPS25 protein shows the evolutionary conservation of the Pro72. (D) The modelling of Pro72Leu (stick representation) mutation on the structure of the human mitoribosome (PDBID: 3J9M) reveals that it is likely to sterically hinder the formation of interprotein contacts with uS17 (yellow) as well as result in a potential destabilisation of the folding of the essential strand-turn-strand in the mS25 (cyan) protein core. The inset of the mitoribosomal structure illustrates the relative position of uS17-mS25 (surface representation) on the small subunit (yellow cartoon); CP, central protuberance.
