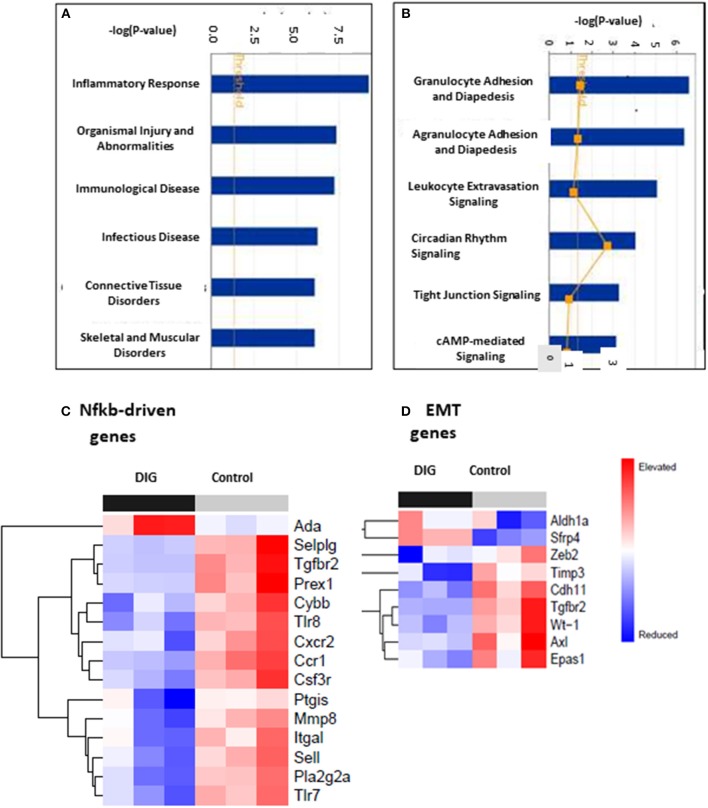
Figure 9.
Gene ontology (GO) and Heat map analysis of digitoxin-dependent genes in stratified primary tumors. (A) Principal Gene Ontology (GO) analysis by IPA of digitoxin effects on transcription profiling of mRNA. The principal drug effects on mRNA and microRNA expression is on Inflammatory Response (#1, P, 2 tailed = ca. 10 −8) and Immunological Disease (#3, P, 2tailed = ca. 10 −6). Other significant and relevant categories include Organismal Injury and Infectious Disease. (B) Secondary Gene Ontology (GO) analysis by IPA of digitoxin effects on Inflammation on transcription profiling of mRNA. Within the category of the “inflammatory response” shown in Part A, the top three sub-GO categories refer to adhesion and diapedesis (movement of cells from blood to tissue) for granulocytes and agranulocytes, and for leukocyte extravasation signaling (all with P, 2 tailed < 1EXP-5). The fourth category is circadian rhythm signaling. (C) Effect of digitoxin on expression of Nfkb-driven mRNAs in stratified primary tumors. Among the top 15 genes in this category, 14, including Tgfbr2, are suppressed by treatment with digitoxin. (D) Effect of digitoxin on expression of Epithelia-to–Mesenchymal (EMT) mRNAs in stratified primary tumors. Of the 9 genes in this category, two are elevated (Aldh1a and Sfrp4) and 7 are reduced (Zeb4, Timp3, Cdh11, Tgfbr2, Wt-1, Axl, and Epas1).