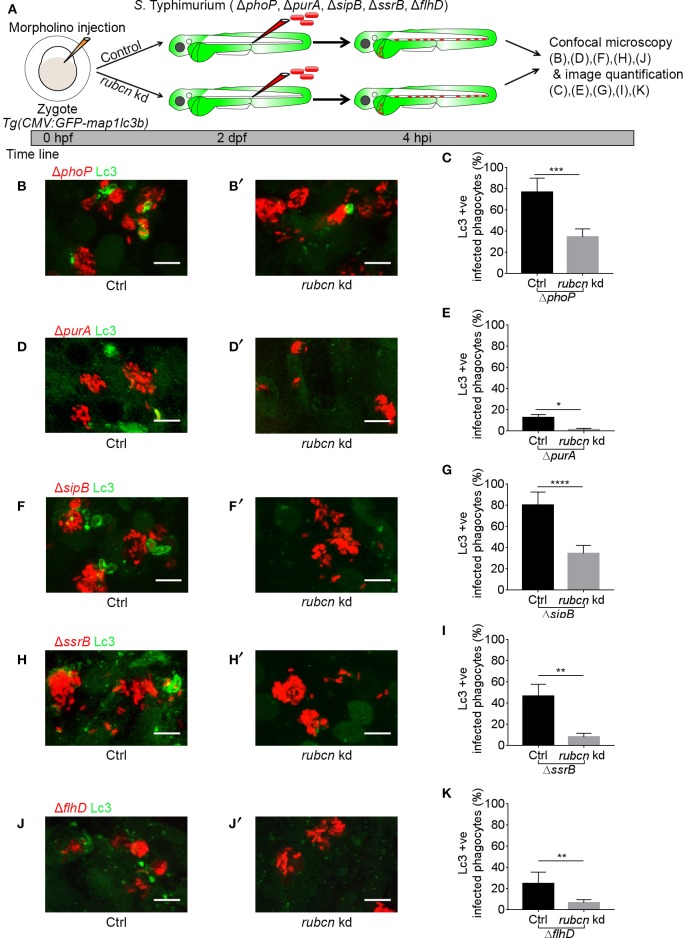
Figure 3.
Host LAP response to S. Typhimurium mutants in Rubicon-depleted embryos. (A) Work flow and time line of experiments in (B–K). (B,D,F,H,J) Representative confocal micrographs of GFP-Lc3 positive infected phagocytes with mCherry-labeled S. Typhimurium mutant strains ΔphoP (B), ΔpurA (D), ΔsipB (F), ΔssrB (H), and ΔflhD (J) in Rubicon-depleted hosts (B',D',F',H',J') along with respective controls (B,D,F,H,J). (C,E,G,I,K) Quantification of GFP-Lc3-Salmonella association for S. Typhimurium mutant strains ΔphoP (C), ΔpurA (E), ΔsipB (G), ΔssrB (I), and ΔflhD (K) in Rubicon-depleted larvae and their respective controls. Numbers of infected phagocytes positive or negative for GFP-Lc3-Salmonella associations were counted from confocal images and the percentages of Lc3-positive over the total were averaged from five embryos per group. Error bars represent the SD. One of the two replicates is shown. Scale bar (B,D,F,H,J) = 10 μm, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.