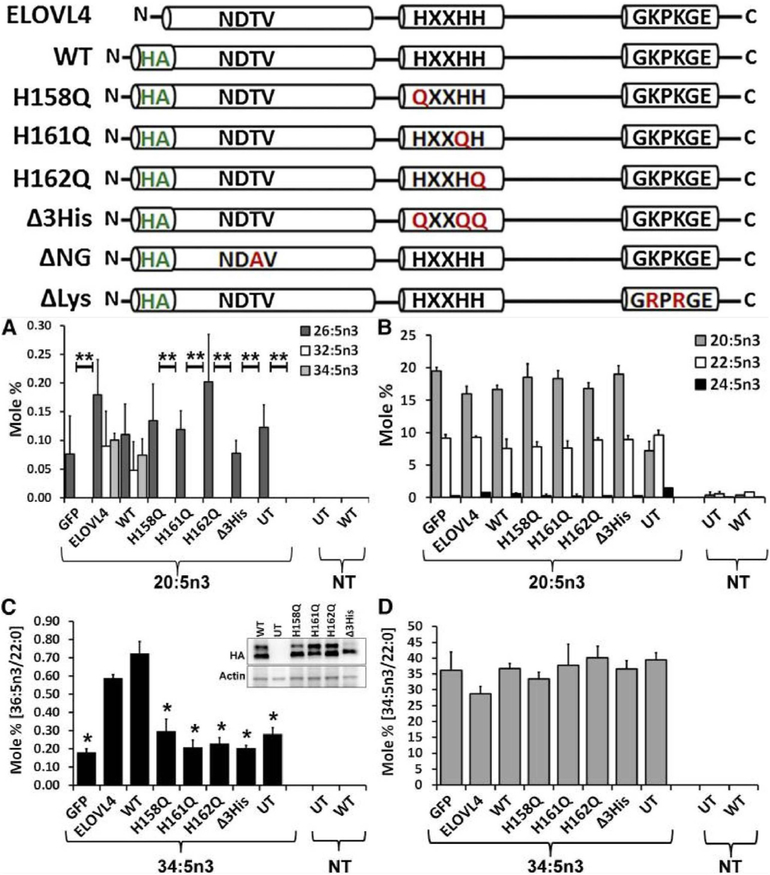
Fig. 5.
ELOVL4 active site mutants are deficient in VLC-PUFA biosynthesis. Schematic representation of untagged (ELOVL4) and HA-ELOVL4 constructs indicating individual mutations in active site (H158Q, H161Q, H162Q, and triple mutant Δ3His), N-glycosylation mutant (T22A; ΔNG), and lysine mutant (K308, 310R; ΔLys). (A) Elongation of 20:5n3 in HEK293T cells to 32:5n3 and 34:5n3 in ELOVL4 and WT, but not in catalytic dead mutants (histidine mutants) or GFP-expressing and UT controls. (B) Relative mole percent of 20:5n3, 22:5n3, and 24:5n3 with and without (NT) supplementation showing comparable levels of these FAs across samples in transduced HEK293T cells. (C) Elongation of 34:5n3 to 36:5n3 normalized to 22:0 in HEK293T cells expressing ELOVL4 and WT, but not in active site mutants, which were comparable to controls (GFP and UT). Data are represented as the mean ± SD (n = 3). Significance was assessed in comparison to WT; *P < 0.05; **P < 0.01. (Inset: adenoviral-mediated expression of HA-ELOVL4 proteins supplemented with either 20:5n3 or 34:5n3). (D) Levels of 34:5n3 internalized across treated and NT samples showing comparable levels of the precursor. Adapted reproduction with permissions from: Logan et al. (2014). J Lipid Res. Apr; 55(4): 698–708. doi:10.1194/jlr.M045443. © 2014 by the American Society for Biochemistry and Molecular Biology, Inc. https://creativecommons.org/licenses/by/3.0/.