Abstract
Fungi encompass, in addition to classically well-studied lineages, an ever-expanding diversity of poorly known lineages including zoosporic chytrid-like parasites. Here, we formally describe Amoeboradix gromovi gen. et sp. nov. comprising a set of closely related strains of chytrid-like parasites of the green-yellow alga Tribonema gayanum unusually endowed with amoeboid zoospores. Morphological and ultrastructural features of A. gromovi observed by light and transmission electron microscopy recall previous descriptions of Rhizophydium anatropum. A. gromovi exhibits one of the longest kinetosomes known in eukaryotes, composed of microtubular singlets or doublets. To carry out molecular phylogenetic analysis and validate the identification of different life cycle stages, we amplified 18S rRNA genes from three A. gromovi strains infecting T. gayanum cultures, single sporangia and single zoospores. Molecular phylogenetic analyses of 18S+28S rRNA concatenated genes of the type strain revealed that A. gromovi is closely related to the recently described species Sanchytrium tribonematis, another parasite of Tribonema that had been tentatively classified within Monoblepharidomycetes. However, our phylogenetic analysis with an extended taxon sampling did not show any particular affinity of Amoeboradix and Sanchytrium with described fungal taxa. Therefore, Amoeboradix gromovi and Sanchytrium tribonematis likely represent a new divergent taxon that remains incertae sedis within Fungi.
Keywords: parasites of algae, ultrastructure, molecular phylogeny, life cycle, amoeboid zoospore, kinetosome
Introduction
In recent years, the generalized use of molecular phylogeny tools to study microbial communities is unveiling an increasingly expanding diversity of microbial eukaryotes. In the particular case of Fungi, the combination of molecular phylogeny and electron microscopy has revealed a wide cryptic diversity of zoosporic fungi. Thus, in the past decade several new orders of chytridiomycetes, some of which had already been predicted based on molecular phylogeny analysis (James et al. 2006), have been formally described (Letcher et al. 2006; Mozley-Standridge et al. 2009; Simmons et al. 2009; Longcore & Simmons 2012; Karpov et al. 2014). At the same time, some organisms traditionally considered typical chytrids dramatically changed their taxonomic classification to match their position in molecular phylogenetic trees. An illustrative example is that of the former chytridiomycete genus Olpidium, which was re-classified as a member of the zygomycete clade (James et al. 2006; Sekimoto et al. 2011).
Some years ago, we presented the chytrid-like strain X-44 CALU, a parasite of the yellow-green alga Tribonema gayanum, as the new genus and species Amoeboradix gromovi (ECOP meeting; Mamkaeva et al. 2007). However, the molecular phylogenetic affiliation of this strain and its formal description remained to be established. A. gromovi exhibited a typical chytrid morphology, similar to that of Rhizophydium anatropum (A. Br.) Karling (Supplementary Fig. S1; Couch 1935; Letcher and Powell 2012), but had amoeboid zoospores with a pseudocilium instead of a flagellum and an extremely long kinetosome composed of microtubular singlets (Mamkaeva et al. 2007). Amoeboid zoospores also contained a lipid-microbody complex, but lacked dictyosomes and fenestrated cisterna. Rounded cysts formed rhizoids inside the host; thick-walled sporangia were normally asymmetrical, rounded to pear-shaped. Unfortunately, the strain X-44 was subsequently lost and, with it, the possibility to obtain sequence data for this strain. Since this preliminary description, we have enriched more strains displaying the same morphological characters as X-44 from different freshwater habitats (X-113, X-114 and X-130).
Here we present a detailed light and electron microscopic study of the newly enriched chytrid-like strains X-113, X-114 and X-130 parasitizing Tribonema gayanum and compare it with the original A. gromovi strain X-44. 18S rRNA gene sequences of the new strains are highly similar. Molecular phylogenetic analyses based on concatenated 18S and 28S rRNA genes of X-113, now considered the type strain, place this lineage together with the recently described Sanchytrium tribonematis (Karpov et al. 2017) as an independent fungal group without clear affinities to known fungal taxa. Based on this information, we now provide a formal diagnosis for the new species and genus Amoeboradix gromovi.
Results
The strains X-44, X-113, X-114 and X-130 of Amoeboradix gromovi have similar stages of the life cycle (Figs 1-8). Zoospores attach to the surface of the parasitized algal filament and encyst. Cysts produce rhizoids that penetrate the host cell and grow to produce an elongated asymmetric sporangium with 1-3 papillae for zoospore discharge. Amoeboid zoospores endowed with a posterior pseudocilium are released from the sporangium and infect new algal cells.
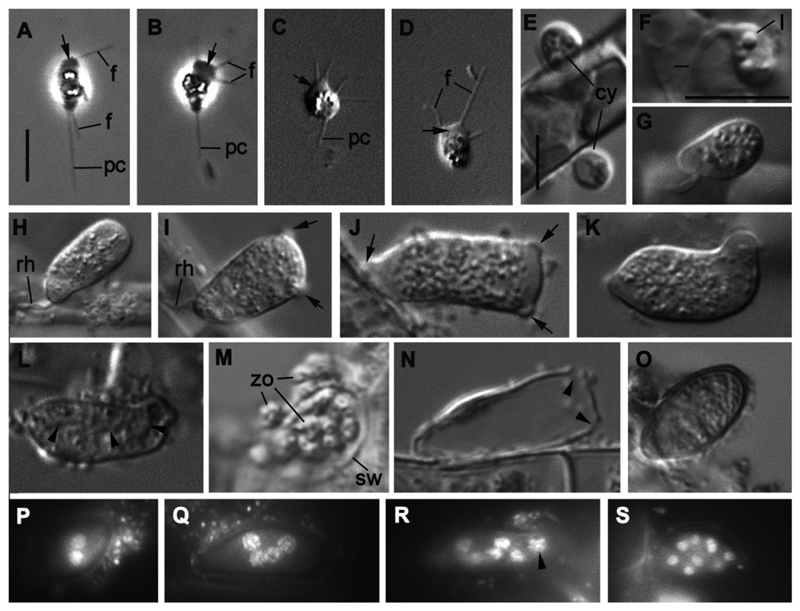
Fig. 1.
Light microscopy of the life cycle stages of strain X-113 on the filaments of Tribonema gayanum. A-D – amoeboid zoospores (in D with retracted pseudocilium), E – two cysts, F-G – young growing sporangium with rhizoid, H-K – mature sporangium of different shape with papillae (arrows), L – sporangium with formed zoospores (arrowheads show the borders between cells), M – zoospore releasing, N – empty sporangium with holes (arrowheads), O – resting spore with thick wall, P-S – DAPI stained nuclei in sporangia from 2 (P) to many (S) (arrowhead shows metaphase plate). Scale bars: A-D - 5 μm; E – 5 μm; F-S - 10 μm.
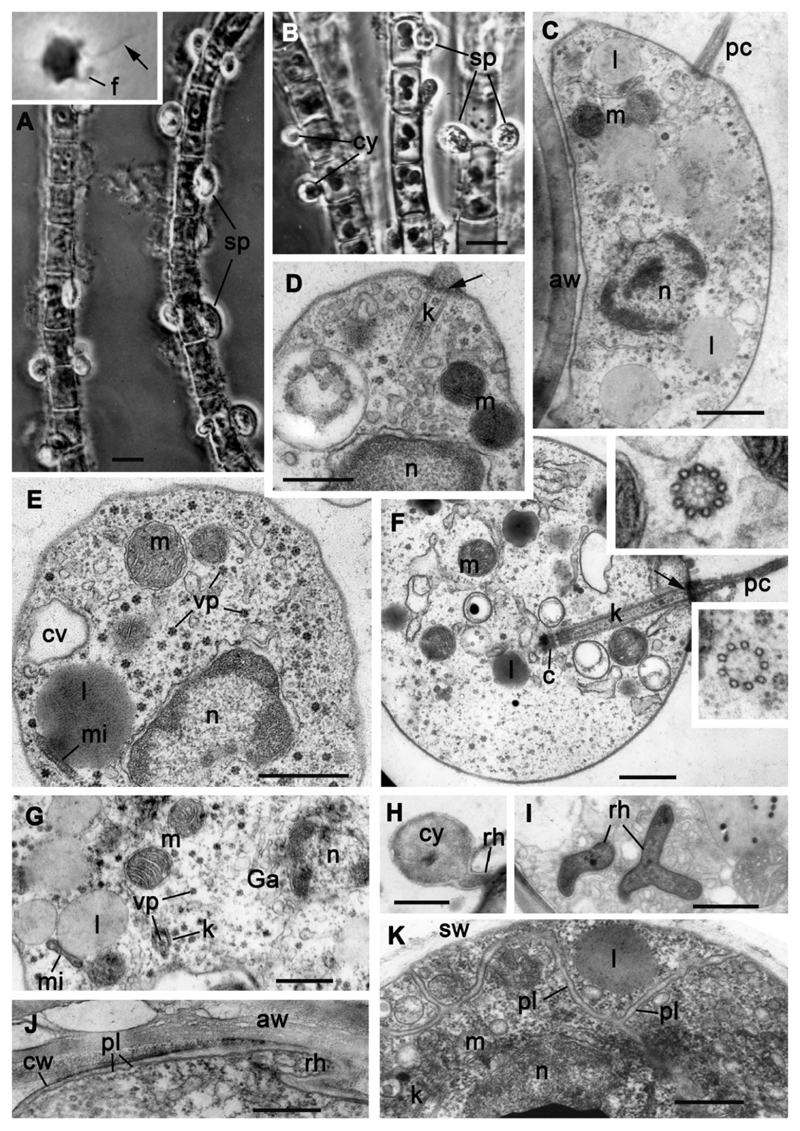
Fig. 8.
Morphology of strain X-44 CALU. A, B – sporangia and cysts on the filaments of Tribonema gayanum, insert in A: amoeboid zoospore with pseudocilium (arrow). C-G – ultrastructure of zoospore. Inserts in F: transversal sections of kinetosome through the hub (upper), and middle (lower). H – cyst with rhizoid, I – branched rhizoid in the host cytoplasm, J – base of rhizoid of the cyst, K – zoospores in sporangium. Arrows in D and F label the septa. Scale bars: A, B – 10 μm; C, D, F, G, I-K – 500 nm; E – 1 μm; H – 2 μm.
A more detailed morphological description follows, starting with the better studied strain X-113, which we now consider as the type.
Light microscopy and ultrastructure of strain X-113
A. gromovi X-113 displays oval to pear-shaped sporangia on filaments of the algal host, T. gayanum (Fig. 1 H-L). The amoeboid zoospores encyst on the algal surface and the cyst then develops prominent rhizoids that penetrate the host cell. These are visible by light microscopy only on the surface of the algal filament (Fig. 1 E-G). Each cyst enlarges and becomes a young sporangium with lipid globules and granular contents. It subsequently grows into a mature oval or pear-shaped asymmetric sporangium producing zoospores (Fig. 1 H-M). The resting spore, or sporocyst, was observed in old cultures. It has ellipsoid shape, thick slightly brownish cell wall, and its dimensions are 6 x 12 μm (Fig. 1 O; Table 1).
Table 1. Comparison of major phenotypic characters between Amoeboradix gromovi strains X-44 and X-113 with Rhizophydium anatropum.
| Amoeboradix gromovi (present paper) |
Rhizophydium anatropum (Braun 1856; Couch 1935) |
||
|---|---|---|---|
| X-44 | X-113 | ||
| Mature sporangium size (μm) and number of papillae | 9 x 16 with 1-2 papillae | 8-10 x 16-18 with 2-3 papillae | 9-14 x 16-25 with a papilla |
| Zoospore size (μm) | 2-3 x 5 | 2-3 x 3-4 | 2.1 x 5 |
| Cyst size (μm) | 5 | 3-4 | 2.2-3 |
| Resting spore size (μm) | - | 6 x 12 | 10 |
| Flagellum/pseudocilium; length (μm) | Pseudocilium; 8 | Pseudocilium; 8 | Flagellum; 10 |
| Rhizoid/haustorium | rhizoid | rhizoid | peg-like haustorium |
| Kinetosome structure; length (μm) | Singlets; 1.5 | Singlets/doublets; 1.8-2.2 | ? |
| Viral particles | + | + | ? |
Zoospore
Amoeboid zoospores, 4-5 μm long and 3 μm wide with few prominent lipid globules, produce thin filopodia at any place of the cell surface during their movement (Fig. 1 A-D). These amoeboid cells normally exhibit a broad hyaline pseudopodium (lamellipodium) at the anterior end that can also form subfilopodia (Fig. 1 A-D). Most zoospores have a tracking posterior pseudocilium of different lengths (up to 8 μm), but this pseudocilium is rather labile (Fig. 1 A-C) and can be totally retracted (Fig. 1 D).
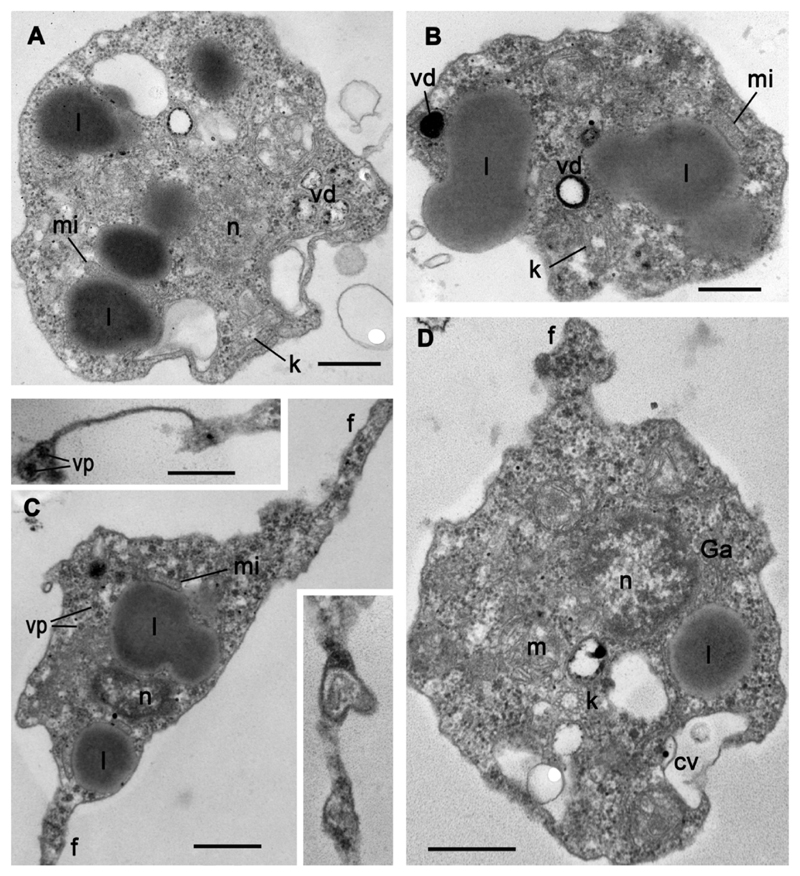
Using serial ultrathin sections, we studied four released zoospores (Fig. 2) and seven zoospores inside three mature sporangia (Figs 3-4). The most prominent organelles and structures of the zoospore are large lipid globules, which seem to fuse in one huge curved rosary chain, and a long kinetosome, 1.8-2.2 μm in length (Fig. 4 A, D). A nucleus in the center of the cell is surrounded by relatively dense cytoplasm with scattered ribosomes. Mitochondria have lamellar cristae, and a small thinly granulated microbody is closely associated with lipid globules forming a microbody-lipid complex (MLC). A Golgi body is present near the nucleus, and a contractile vacuole at the cell periphery (Fig. 2 A, D). Some vesicles ca. 200-300 nm in diameter contain dark material at different degrees of degradation (Fig. 2 A, B, D). As they are associated with lipid globules (Fig. 2 A), we suggest that these vesicles can take part in lipid utilization and refer to them as degradation vesicles (dv). Several profiles of filopodia filled with homogeneous cytoplasm are present around the cell body (Fig. 2 C). Star-shaped structures of about 50 nm in diameter are present throughout the cytoplasm of the parasite (Fig. 2 C). They usually consist of 6 globular units. They are larger than ribosomes and do not seem to be ribosomal clusters. Because of their regular shape and dimension, they might well be viral particles or, perhaps, rosettes of glycogen particles. These particles are present in the cytoplasm and filopodia of free zoospores (Fig. 2 C) but are not obvious in the intrasporangial zoospores (Figs 3, 4).
Fig. 2.
Ultrastructure of four different released zoospores of strain X-113. Inserts in C show more distal parts of filopodia. Scale bars: A-D – 500 nm; both inserts – 250 nm.
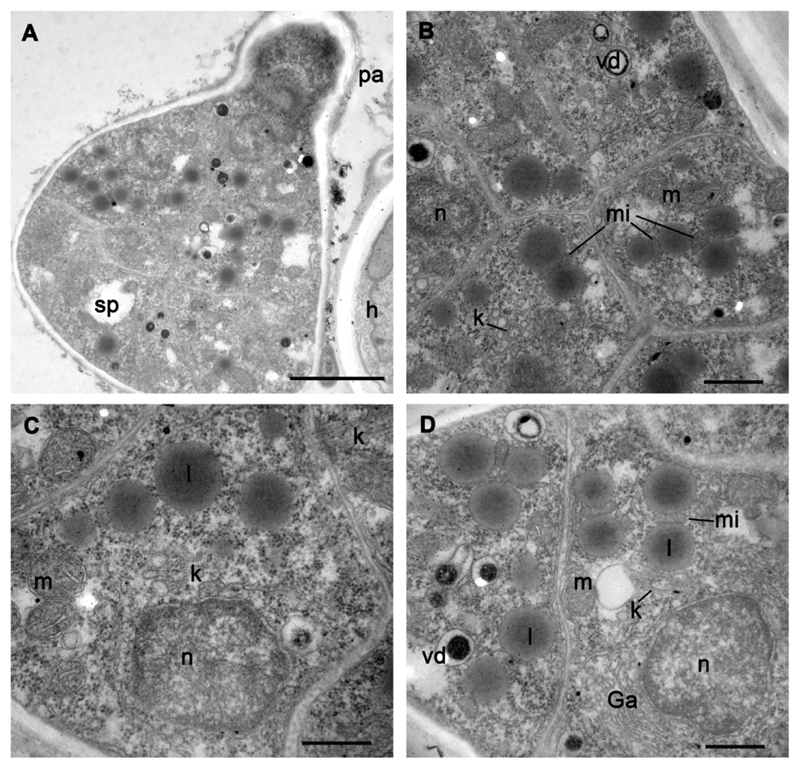
Fig. 3.
Ultrastructure of sporangium with fully formed zoospores (A) and zoospores inside sporangium (B-D) of strain X-113. A – sporangium with papilla, B-D – zoospores from three different sporangia. Scale bars: A - 2 μm; B-D – 500 nm.
Fig. 4.
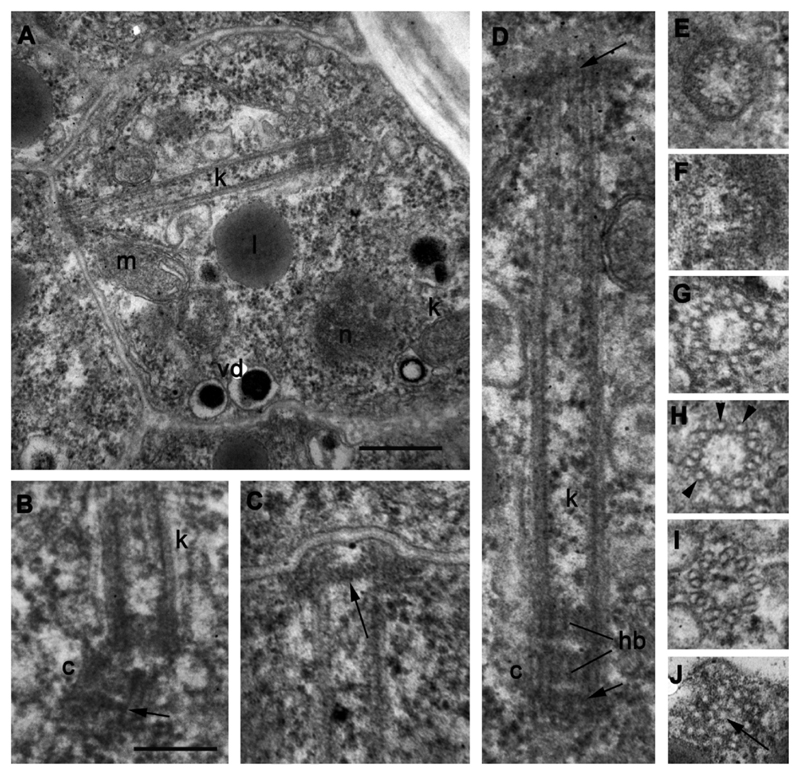
Kinetosome and centriole structure in mature zoospores of strain X-113 inside sporangium. A – longitudinal section of kinetosome and centriole in the cell, B – centriole at blunt angle to kinetosome (arrowhead shows distal septa at the distal end of centriole), C – distal end of kinetosome with transversal septa (arrow) in sporangial cell, D – enlarged kinetosome/centriole complex from A showing their antiparallel position and transversal septae (arrows), E-J – transversal sections of kinetosome at different levels from distal tip (E, F – consecutive sections), to the middle (G-I), and proximal end (J) with hub (arrow). Arrowheads on H mark microtubular singlets. Scale bars: A – 400 nm; B-J – 200 nm.
The mature sporangium contains roughly 12-16 zoospores, which are generally the same as in released zoospores (compare Fig. 2 with Figs 3, 4 A). An endomembrane system and mitochondria with several cristae and a dense matrix are well developed in sporangial zoospores (Fig. 3 D), showing active cell proliferation. The kinetosome is still in the vicinity of the nucleus although later, in released zoospores, the kinetosome may migrate to the cell periphery (Fig. 2 A, B). The mature zoosporangium has papillae (normally 2-3) that become exit pores for zoospore discharge: empty sporangia have holes where papillae lay (Fig. 1 N) and the papillae are also found in ultrathin sections of sporangia with mature zoospores (Fig. 3 A).
The kinetosome in mature zoospores is connected with a short centriole near the nucleus (Fig. 4 A, B, D). The kinetosome grows on the base of the centriole from its proximal end, being antiparallel to the latter on the same longitudinal axis (Fig. 4 A, D). Both structures have a hub at their proximal end. The kinetosome can be detached from the centriole and locates at a blunt angle to it (Fig. 4 B). The distal end of the kinetosome is anchored to the plasma membrane and has a partition somewhat similar to the transversal plate and transitional fibers (Fig. 4 C, D). The kinetosome is composed of 9 microtubular doublets at the proximal 1/3 part, which is 190-200 nm wide, and of 9 microtubular singlets at the middle and distal parts, which are 160-170 nm wide (Fig. 4 E-J).
Cyst
After encystment, the cell has the same diameter as the rounded zoospore. The cyst wall is composed of a thin electron-opaque outer layer (ca. 15-20 nm) spaced from the plasma membrane by approximately 100 nm (although this could be due to a fixation artefact) (Fig. 5 A, B). The outer part of the rhizoid is ca. 5-8 μm long, and 0.5-0.65 μm thick (Table 1; Fig. 1 F-I). It can be straight or curved depending on the place of encystment on the surface of the algal cell. The cyst wall covers the rhizoid, which normally enters the host in between the inner and outer half of the Tribonema cell wall (not shown). Rhizoids are not visible inside the host cell by light microscopy. Transmission electron microscopy shows that the rhizoid contains cytoplasm with cytoplasmic microtubules and vesicles (Fig. 5 B). The cyst has a large nucleus with prominent nucleolus and uncondensed chromatin (Fig. 5 B). Several large lipid globules are associated with the microbody, and mitochondria have flat cristae (Fig. 5 A, B). Small dictyosomes of the Golgi body can also be found in the cyst. Two antiparallel centrioles are visible in one section (Fig. 5 B).
Fig. 5.
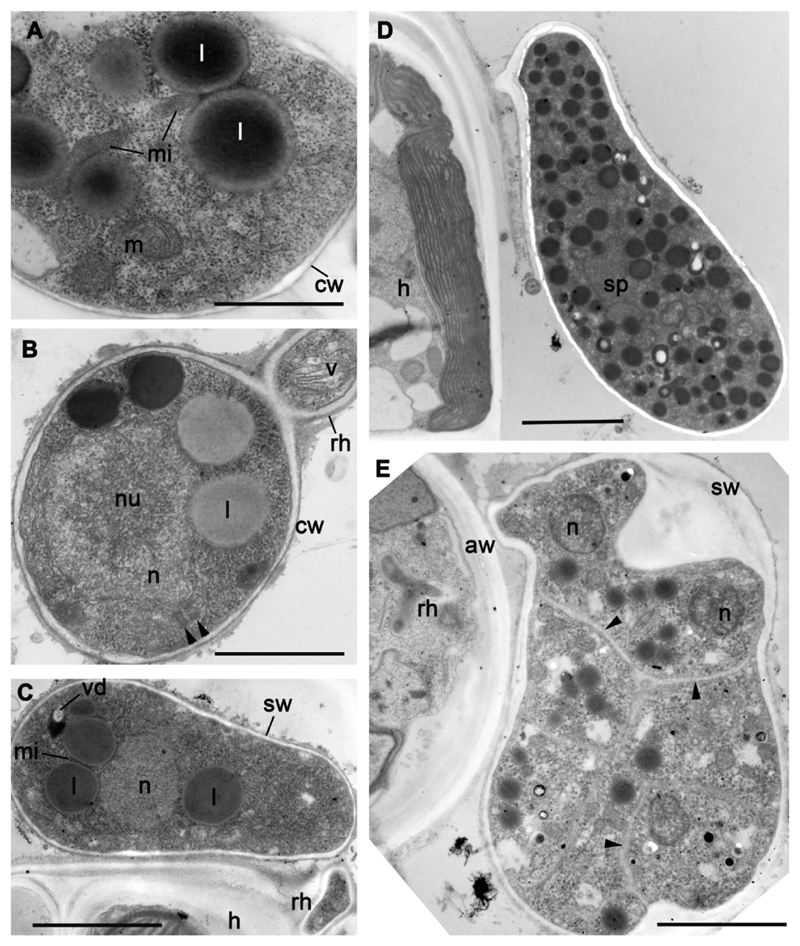
Ultrastructure of cysts (A, B) and sporangia (C-E) of strain X-113. A – microbody-lipid globule complex, B – cyst with rhizoid (arrowheads show two short centrioles), C – young sporangium, D – mature sporangium with several nuclei, E – sporangium with fully formed zoospores (arrowheads show borders between the cells). Scale bars: A – 1 μm; B – 2 μm; C - 3 μm; D-E – 4 μm.
Sporangium
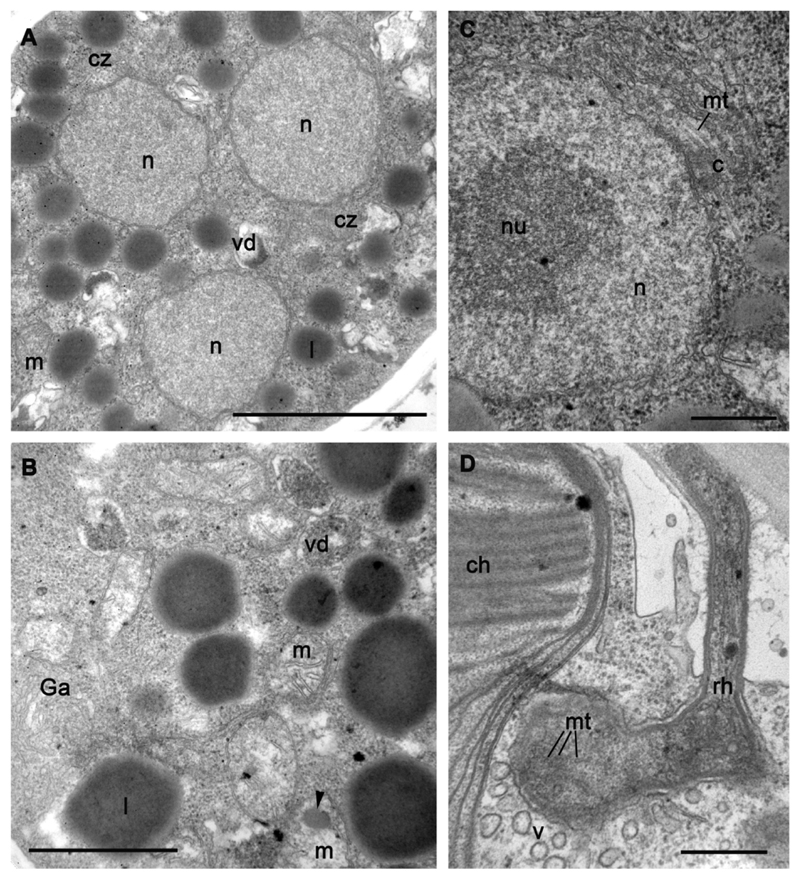
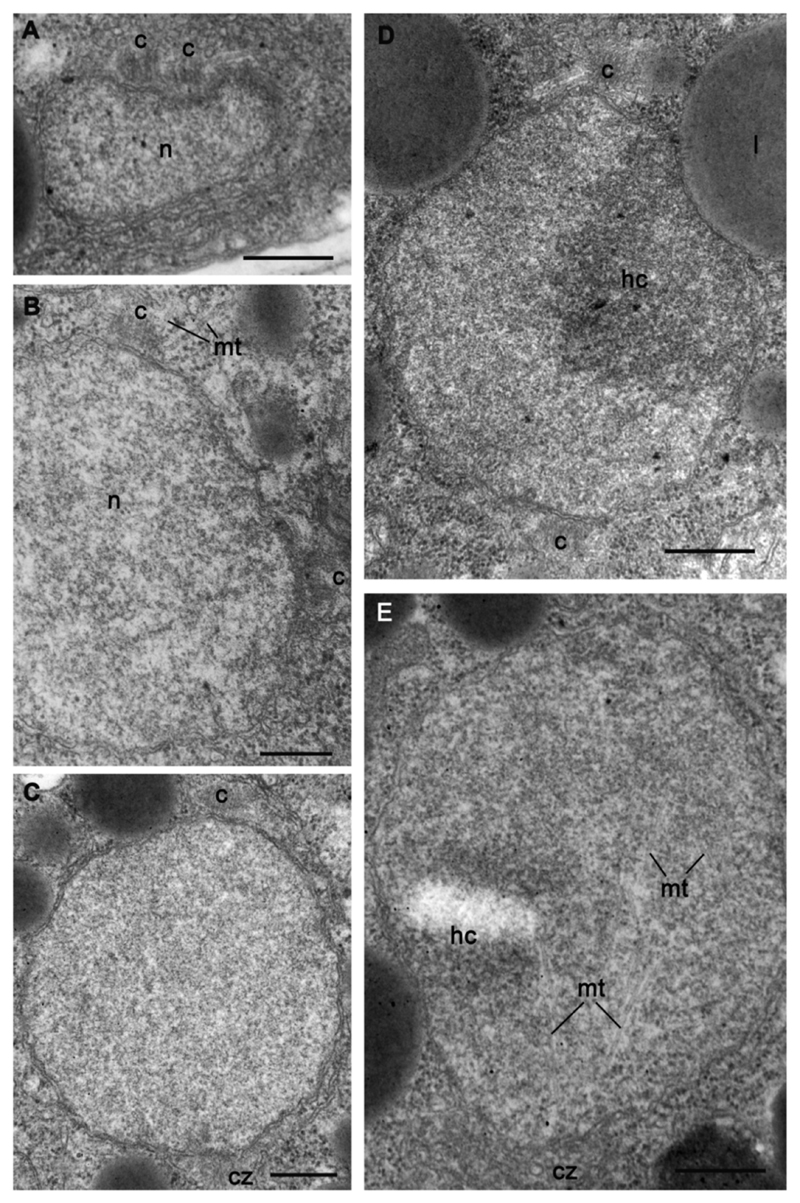
The immature sporangium has several nuclei, which are not visible with DIC microscopy observation of living material, but can be easily counted with DAPI staining (Fig. 2 P-S). Unexpectedly, the sporangium dimension is not directly proportional to the number of nuclei: sporangia of nearly the same dimensions have different numbers of nuclei, which may also have variable diameters (Fig. 2 P-S). A metaphase plate can be noted in some nuclei (Fig. 2 R) showing their asynchronous division. In ultrathin sections, we normally found 2-3 nuclei with eccentric nucleoli (or heterochromatin) and uncondensed chromatin (Figs 5 C-E, 6, 7). Small centrioles with radiating microtubules are found “sitting” at the nuclear envelope (Figs 6 C, 7).
Fig. 6.
Ultrastructure of sporangium (A-C) and rhizoid (D) of strain X-113. A – nuclei in the middle of sporangium, B – portion of cytoplasm (arrowhed shows amorphous inclusion in mitochondrium), C – interphase nucleus with centriole, D – rhizoid structure. Scale bars: A – 2 μm; B – 500 nm; C – 500 nm; D – 1 μm.
Fig. 7.
Dividing nuclei in sporangium of strain X-113. A-B – separation and migration of centrioles in prophase, C-E – nuclei at metaphase with internal spindle (E). Scale bars: A-E – 500 nm.
The cell cytoplasm is filled with numerous lipid droplets, mainly spherical (Fig. 5 D). Small vacuoles with amorphous contents often occur in the cytoplasm, being associated with lipid globules in some sections (Fig. 5 C, 6 A, B). Small microbodies are also present in contact with lipid globules as in the zoospores, being clamped between the globules. Numerous mitochondria with predominantly flat cristae spread throughout the sporangium. Some mitochondria have dark inclusions of unknown nature in their matrix (Fig. 6 B). The mature sporangium has the shape of an asymmetric pear of ca. 16-18 x 8-10 μm and lays on the algal filament at an angle of 45-60° with respect to the longitudinal algal filament axis. The sporangial wall is much thicker than the cyst wall; it is composed of a thick inner electron translucent layer and a thin electron dense outer layer. Its total thickness may reach up to 300 nm.
Nuclear division
Different stages of dividing nuclei can be seen in the sporangia (Fig. 7). The classical image of two orthogonal centrioles at the interphase nucleus has not been observed. At the first stage of nuclear division, a new centriole appears parallel to the mother one (Fig. 7 A). Then both centrioles migrate to the nuclear poles producing microtubules (Fig. 7 B-E). The nucleus is normally electron-translucent at these stages, exhibiting uncondensed chromatin and profiles of disordered intranuclear microtubules. However, in some sections, perhaps illustrating the prometaphase or metaphase, chromatin partly condensates and the microtubules form an intranuclear spindle. The nuclear envelope seems to stay intact and centrioles with radiating microtubules localize at the opposite nuclear poles (Fig. 7 E). The anaphase and telophase were not observed.
The ultrastructure of the strains X-114 and X-130 was not studied in detail, but the light microscopic observations did not revealed morphological differences between X-113, X-114 and X-130.
Strain X-44
All life cycle stages of this strain are similar to those of A. gromovi strains X-113, X-114 and X-130. When attached to the algal filaments in infected cultures, X-44 strain forms numerous, small and round cysts, and oval or pear-shaped often asymmetrical sporangia up to 16 μm length, and 9 μm width, with granular content (Table 1; Fig. 8 A, B). A thin rhizoid penetrates the algal cell from the base of the sporangium. The wall of the empty sporangium is thick and contains an apical or subapical cylindrical papilla 2.5-3.0 x 2.5-3.0 μm. The motile stage of the parasite is represented by small amoeboid zoospores, oval, or with several filopodia. Amoeboid zoospores are about 2-3 μm in diameter, up to 5 μm long during crawling movement (Fig. 8 A insert). They have a posterior pseudocilium, up to 8 μm long. Amoeboid zoospores attach to the algal cell wall and transform into the oval cysts of ca. 5 μm in diameter (Fig. 8 A, B). A rhizoid appears from the base of the cyst and penetrates the algal cell wall to enter into the host cell. The round, thin-walled cyst transforms into the oval thick-walled sporangium.
Amoeboid zoospores contain a central nucleus, mitochondria with lamellar cristae, contractile vacuole complex and lipid globules associated with small microbodies (Fig. 8 C-G). The ribosomes and numerous virus-like particles ca. 50 nm in diameter are scattered throughout the cytoplasm. An endoplasmic reticulum connected with the nuclear envelope is present, but a Golgi apparatus with stacked cisternae has not been observed (Fig. 8 G). The pseudocilium has a long kinetosome composed of 9 single microtubules 160-170 nm wide and 1.5 μm long (Fig. 8 D, F). The kinetosome contains a hub at its proximal end and transversal septa at the distal end. In some sections, the kinetosome is connected to a small centriole (Fig. 8 F).
Since this strain was lost, the structure of cysts and sporangia could not be investigated in detail. In available X-44 preparations, cysts appeared covered by a thin cyst wall and penetrate the host cell wall with rhizoids that can branch in the host cytoplasm (Fig. 8 H-J). Mature zoospores in the sporangium have all the structures and organelles found in released zoospores (Fig. 8 K).
Growth characteristics
After inoculation with infected algae, cultures of T. gayanum die after 2-6 weeks depending on the inoculum size. The parasite develops in the dark or under illumination, but only on live algae. Good growth of the parasite is observed on Tribonema gayanum (initial host), T. vulgare Pasch. CALU 1166 and Ulothrix tenerrima Kutz. CALU 1167. The parasite does not develop in cultures of the following algae: Chlorococcum (12 strains examined), Ankistrodesmus (9 strains), Dictiosphaerium (4 strains), Bracteacoccus (3 strains), Kirshneriella (2 strains), Pediastrum, Chlorotetraedron, Spirogira sp., Stigeoclonium attenuatum, Ulothrix limnetica, and Trentepohlia uncinata. The parasite does not invade threads of algae growing on solid media and, therefore, we could not obtain discrete plaques of parasite on algal lawns growing on the surface of solid media. In cultures inoculated with infected algae, algal death was observed only when parasite sporangia appeared on the algal threads. Cultures destroyed by the parasite remain infectious for a long time, even after one year of storage in the refrigerator.
Molecular phylogeny
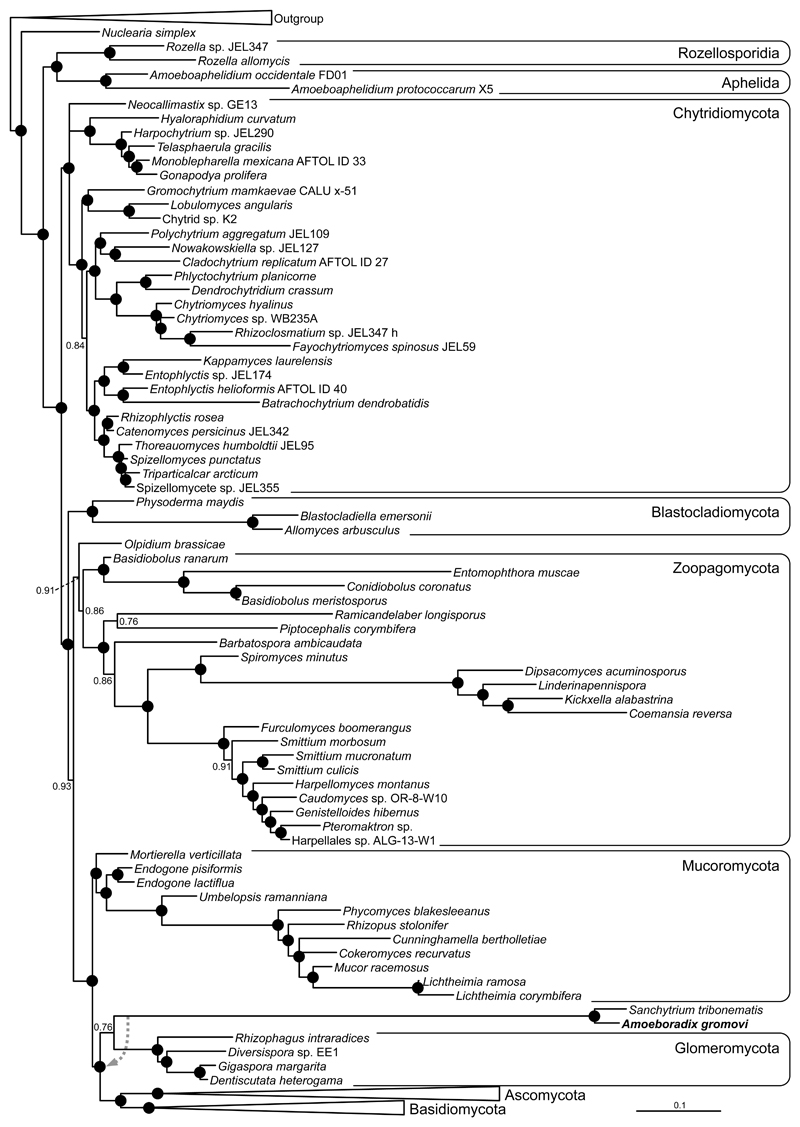
We collected sporangia from strains X-113, X-114 and X-130 and individual amoeboid zoospores by micromanipulation from strain X-113 for DNA extraction and 18S rRNA gene PCR amplification and sequencing. The 18S rRNA gene sequences from all these strains are >99.5% similar to each other, suggesting that they correspond to different strains of the same species. Phylogenetic analysis of these sequences together with other eukaryotic homologues confirmed the fungal affinity of Amoeboradix although with poor statistical support (Supplementary Fig. S2). This was consistent with the strains’ low sequence similarity to the closest fungal sequences available at the time (less than 85%). Consequently, we also amplified and sequenced the 28S rRNA gene (as a single contig 18S + 5.8S + 28S rRNA gene cluster) to increase the phylogenetic signal. The 28S rRNA gene exhibited an equally low similarity with most available fungal sequences in GenBank (e.g. 83% sequence identity shared with one of its closest BLAST hits, Cryptococcus neoformans). However, both 18S and 28S rRNA gene sequences were highly similar (ca. 95% identity, excluding intron regions) to those of the recently described Sanchytrium tribonemae, a morphologically similar parasite of Tribonema algae that was tentatively attributed to the Monoblepharidomycetes based on a relatively limited fungal taxon sampling (Karpov et al. 2017). Compared to Sanchytrium, whose rRNA genes contain numerous introns, Amoeboradix is much less intronized. The Bayesian combined 18S+28S rRNA gene phylogenetic tree of Amoeboradix and Sanchytrium including an extended fungal taxon sampling does not support their relationship with Monoblepharidomycetes and fails to retrieve any clear affinity with known fungal clades (Fig. 9). The Amoeboradix-Sanchytrium clade appears somewhat closer to Glomeromycota and Dikarya (Fig. 9), forming a tripartite cluster with a posterior probability of 1, but the relationship of the Amoeboradix-Sanchytrium clade with Glomeromycota lacks any solid support (0.76 posterior probability). In fact, in some of the trees explored during the Bayesian reconstruction, Amoeboradix-Sanchytrium branched at the base of a group composed of Glomeromycota+Dikarya (Fig. 9). Therefore, Amoeboradix and Sanchytrium appear to define a new divergent clade of fungi of unclear affinity. Both species share a very long branch in the 18S+28S rRNA gene phylogenetic tree, such that their unstable position might in principle be due, at least partially, to a long branch attraction artefact (LBA). However, they do not branch close to any of the other long branches of the tree (the Mucorales within the Mucoromycota and some of the Kickxellales species within the Zoopagomycota), suggesting that the position of Amoeboradix and Sanchytrium is not due to LBA. Phylogenomic analysis using many more markers from genome or transcriptome sequence data will be necessary to resolve the phylogenetic position of Amoeboradix and Sanchytrium within the fungal tree.
Fig. 9.
Bayesian phylogenetic tree of fungi including Amoeboradix gromovi and Sanchytrium tribonematis. The tree is based on concatenated 18S+28S rRNA sequences (3761 conserved sites) and is rooted on 12 opisthokont sequences (for the complete tree, see the Supplementary Fig. S3). The dashed arrow on the branch of Amoeboradix+Sanchytrium indicates the alternative position found for this clade in some of the trees explored during the Bayesian analysis. Numbers at nodes are Bayesian posterior probability values, black circles indicate posterior probabilities of 1.
Discussion
With the aim of bringing new light into the phylogeny and taxonomy of poorly studied unusual zoosporic fungi, we present here a formal description and molecular phylogenetic analysis for an atypical set of strains with amoeboid zoospores and one of the longest kinetosomes known to date in eukaryotic cells, which might have been originally observed in the mid nineteenth century. These strains define the new species Amoeboradix gromovi and are closely related to the recently described Sanchytrium tribonematis (Karpov et al 2017). Yet, the phylogenetic position of these two genera remains uncertain, and these organisms might well represent a new fungal clade, reinforcing the idea that new fungal taxa at the level of order or even higher taxonomic rank remain to be properly identified and described in natural environments.
Morphology
The two investigated strains (the lost strain X-44, from preserved material, and the newly isolated strain X-113, now considered the type strain) have similar zoospore structure (including a peculiar kinetosome). The similarity observed in terms of amoeboid zoospores with pseudocilium, sporangia, cyst structure and dimensions (Table 1), mode of nutrition and their preference for the same host also support that these strains belong at least to the same genus. Nonetheless, some minor differences between these strains can be observed: the kinetosome of X-44 zoospores are made of singlets, while that of X-113 zoospores is longer and also contains doublets; the microbody in X-113 is normally positioned in between the lipid globules at nearly all stages of the life cycle, while in X-44 the microbody is more or less free of lipids; the shape of sporangia in X-44 is a more regular ellipsoid with one or two papillae, in contrast with the rather anatropous sporangia with 2-3 papillae found in X-113. Unfortunately, we were only able to obtain rRNA gene sequence data from X-113 and related X-114 and X-130 strains, as strain X-44 was lost some time ago. Thus, it is not possible to estimate their evolutionary distance and their respective positions in molecular phylogenetic trees. Based on zoospore structure, which contain primary characters for chytrid taxonomy (Barr 1978; Powell 2016a), these two strains are similar to each other but in the absence of strain X-44 sequence data, we cannot decide whether the observed strain differences are inter- or intraspecific. Thus, at this stage we propose to describe strains X-44, X-113, X-114 and X-130 under the same genus and species name, Amoeboradix gromovi gen. nov., sp. nov. Future re-isolation and gene sequencing of strain X-44 members might lead to an eventual re-classification of this strain out of this species if the molecular phylogenetic analysis shows significant divergence.
Among zoosporic fungi, several species with amoeboid zoospores have been described within the Blastocladiomycota (Letcher et al. 2016; Powell 2016b; Sparrow 1960; Strittmatter et al. 2015). They produce filopodia and an anterior lamellipodium during their movement, and one of them (Paraphysoderma sedebokerense) retains a posterior flagellum (Strittmatter et al. 2015). They have complex life cycle including sexual and asexual phases, where the amoeboid forms are the infective agents in the vegetative cycle, and the flagellated forms are, probably, gametes that can transform into amoeboid cells for subsequent gamete fusion. The ultrastructure of P. sedebokerense amoeboid zoospores does not reveal long kinetosomes (Letcher et al. 2016). In fact, no report on such long and, at the same time, reduced kinetosome has been published for zoosporic fungi.
Kinetosome structure
Despite the remarkable variety of microtubular sets observed in the flagellar axonemes (Karpov 1987; Seravin 1985), the kinetosomes or basal bodies are more conservative and normally contain 9 triplets of microtubules. Microtubular reduction in kinetosomes is rare. There are few examples known in protists where doublets are found instead of triplets: the pelobiontid Mastigamoeba schizophrenia (Simpson et al. 1997), the bicosoecid Siluania monomastiga (Karpov et al. 1998), the zoospore of Gromochytrium mamkaevae (Gromochytriales, Chytridiomycetes) (Karpov et al., 2014), the male gamete of gregarine Stylocephalus; and the singlets in the kinetosome of male gamete of the coccidian Eimeria necatrix (Schrevel et al. 2013). An extreme case may be represented by the pseudocilium of the zoospores of the aphelid Amoeboaphelidium protococcarum, which seem to lack a kinetosome at its base, as it was not found in the cytoplasm of this species (Karpov et al. 2013). Although clearly visible, the kinetosome of Amoeboradix is highly unusual: it is not only of exceptional size, but it is composed of microtubular singlets in X-44 and of singlets/doublets in X-113.
Usually, the normal length of kinetosomes is approximately of 500 nm, although a few protists have exceptionally long kinetosomes. This is the case of the plasmodiophorid Polymyxa graminis zoospores (Barr and Allan 1982) or the hypermastigine Trichonympha, which holds the record for the longest kinetosomes known (up to 4.5 μm long) (Guichard et al. 2013). However, in all these examples, microtubular triplets compose the kinetosome. Amoeboradix possesses one of the longest kinetosomes known (2.2 μm in length). Yet, despite its extraordinary length, it is a structurally reduced kinetosome, being composed of microtubule singlets/doublets, instead of triplets, and giving rise to a reduced flagellum, an immotile pseudocilium containing a few disordered microtubules. This is an extremely unusual feature in eukaryotic cell biology.
Molecular phylogeny
Molecular phylogenetic trees of 18S rRNA gene sequences obtained from independent sources (infected cultures and single-picked zoospores) for three similar A. gromovi strains (X-113, X-114, X-130) showed that the three are closely related, having almost identical gene sequences (Supplementary Fig. S2). To improve the phylogenetic resolution, we sequenced the 28S rRNA gene and carried out more in-depth phylogenetic analyses using a concatenated 18S+28S rRNA dataset. Amoeboradix turned out to be closely related to Sanchytrium, a recently described genus that was tentatively classified within the Monoblepharidomycetes based on overall morphological resemblance and phylogenetic analysis that included a limited fungal taxon sampling (Karpov et al, 2017). However, our phylogenetic analyses including Amoeboradix and Sanchytrium (Sanchytriaceae) with a wider spectrum of fungal sequences clearly shows that the two genera form a robust monophyletic clade that seems to branch in a basal position to the Glomeromycota and Dikarya cluster (Fig. 9). The Amoeboradix-Sanchytrium clade is very divergent, having an unstable position within the tripartite Sanchytriaceae-Glomeromycota-Dikarya cluster. This phylogenetic position supports the idea that Amoeboradix and Sanchytrium represent a new fungal lineage. Future multi-gene analyses from genome or transcriptome sequences should help to find a fungal home for these strains within established orders or, else, lead to the erection of a novel high-rank fungal taxon (at the level of order or above). For the time being, this lineage should remain incertae sedis within Fungi.
Taxonomy
Because the formal diagnosis of strain X-44 (Mamkaeva et al. 2007) was lacking, the name Amoeboradix gromovi was not valid and registered in international databases. We therefore describe here the type species of Amoeboradix as A. gromovi for strains X-44 and X-113 (see below).
The life cycle stages and the type of nutrition of strain X-113 provides evidence for its fungal affinity. Superficially, it resembles epibiotic chytrids, which produce a branched rhizoidal system for feeding on the host. Many epibiotic monocentric chytridiomycetes that parasitize algae have been described, but nearly all of them have opisthokont zoospores (Sparrow 1960). Some of them demonstrate an amoeboid activity, but have a posterior flagellum for swimming (Powell 2016b). Of course, many unusual chytrids (some with amoeboid zoospores) have been described since the middle of the 19th century by Zopf, Braun, Fischer and others, but most of them were not studied in detail and still have uncertain affinity. One such strange organism, Rhizophydium (originally Chytridium) anatropum (Braun) Karling 1977, has sporangia with the same characteristic shape as our Amoeboradix strains (Braun, 1856): elongated nearly pear-shaped, slightly curved with rounded distal end and attached to the host alga Chaetophora elegans near the proximal pointed end. However, the sporangia of Ch. anatropum are much bigger: 13-14 x 25-33 μm, sometimes 50 μm long, vs 9x18 μm in strains X-44 and X-113 (Table 1). Ch. anatropum has amoeboid zoospores of about 3.3 μm in diameter with a long flagellum of ca. 10 μm, which were unfortunately not illustrated (Braun, 1856). Ch. anatropum produced nearly ovoid resting spores with a thick slightly brownish wall.
Later, Sparrow (1960) wrote that Ch. anatropum zoospores have a posterior flagellum, and enlarged sporangium dimensions (5-14 μm wide by 15-33 μm (rarely 50 μm) long), but without providing an illustration of the zoospores. The most completely illustrated description (reproduced in Fig. S1) of Phlyctidium anatropum (A. Br.) Sparrow was presented by Couch (1935). This description fairly illustrates the characters of our strains. The amoeboid appearance and movement of zoospores were described precisely in particular showing the hyaline anterior zone producing filopodia. The shape and dimensions of sporangia with hyaline papillae, the resting spores, zoospores and cysts are nearly the same (Table 1). We can notice just a few differences: zoospores of P. anatropum have a rod-like flagellum (not a thin pseudocilium), which is longer than the pseudocilium in our strains; sporangia in our strains are smaller and have 2-3 papillae instead of one (P. anatropum); and the rhizoid of sporangia in our strains is longer than the peg-like haustorium of P. anatropum.
These differences might be intraspecific and, as such, not essential for a new species description, but they might also represent more important differences (justifying different genera or even higher-order taxonomic groups). Indeed, Phlyctidium anatropum was re-described later as Rhizophydium anatropum, showing similarities with Rhizophydium spp., and was moved to the order Rhizophydiales (Karling 1977). Unfortunately, the absence of sequence data for those strains precludes their inclusion in phylogenetic trees and therefore evaluating their true evolutionary relatedness with our strains. Nevertheless, according to our molecular data, our strains are extremely distant from other known fungal species or genera and do not show any robust phylogenetic affinity with those orders. We therefore have decided to place this taxon among incertae sedis within Fungi. The zoospore structure with such unusual long and reduced kinetosome with a pseudocilium evidences the uniqueness of this organism among known zoosporic fungi. Therefore, we are providing here a formal description of the genus Amoeboradix with X-113 being the type strain for the species.
Amoeboradix Karpov, López-García, Mamkaeva et Moreira gen. nov.
Zoosporic fungus with monocentric, epibiotic sporangia and amoeboid zoospores having posterior pseudocilium that emerges from long kinetosome composed of microtubular singlets or doublets.
Etymology: amoeba L – amoeboid zoospores, radix L – root, producing rhizoid when encysted.
Type species A. gromovi Karpov, López-García, Mamkaeva et Moreira sp. nov.
A. gromovi Karpov, López-García, Mamkaeva et Moreira sp. nov.
Amoeboid zoospores of 2-3 x 3-4 μm with posterior pseudocilium up to 8 μm in length, which can be totally reduced; length of kinetosome is ca. 2 μm. Sporangia of variable shape from pear-shaped with rounded broad distal end and pointed proximal end to curved asymmetrical sac-like 8-10 μm width 16-18 μm long with prominent rhizoid and 2-3 papillae for zoospore discharge; spherical cysts about 3-4 μm in diameter; resting spore is ovate thick walled 12x6 μm. Parasite of Tribonema gayanum, T. vulgare and Ulothrix tenerrima.
Etymology: in honor to Boris Gromov, who investigated the strain X-44 of this species.
Type: Fig. 1 in Karpov et al., present paper. Strain X-113 (CCPP ZIN RAS) isolated by M.A. Mamkaeva from sample X-113 in 2014; roadside ditch in the vicinity of Sergievka park, Stary Petershof, St. Petersburg, Russia. The complete cluster including 18S, 5.8S and 28S rRNA gene sequences of strain X-113 is deposited in GenBank with accession number MG256970.
The genus Sanchytrium is closely related to Amoeboradix and has smaller and more round sporangium (appr. 10 μm diam.) with one to three papillae and the zoospores of somewhat similar dimensions (about 3 μm when rounded) with rod-like flagellum (perhaps, a pseudocilium, as we observed the attached zoospore) 5 μm in length (Karpov et al. 2017). There is no ultrastructural data on S. tribonematis to compare the cell interior and estimate the kinetosome structure in this species. Both genera belong to the family Sanchytriaceae Karpov et Aleoshin 2017 which is Fungi incertae sedis.
Material and Methods
Isolation and cultivation of Amoeboradix gromovi gen. et sp. nov.
The morphological data on the strain X-44 CALU (Collection of Algal Cultures in St. Petersburg State University) were obtained from preserved material produced in the Gromov’s lab earlier. The strain X-44 was isolated from the water of a sand-pit in Lomonosov district, Leningrad region, Russia, before being subsequently lost. Tribonema gayanum Pascher CALU-20 growing in N 1 medium (Gromov and Titova 1991) was inoculated with a sample of water filtered through a piece of gauze. After several weeks of incubation the culture lost its green color and threads of empty cell walls covered with characteristic sporangia were observed. A subculture was obtained by inoculation of axenic Tribonema cultures growing in liquid medium with a single infected filament collected from an infected culture via capillary micromanipulation. Algae used in inoculation experiments with X-44 included strains from the CALU collection (Gromov and Titova 1991). Strains X-113, X-114 and X-130 were isolated by M.A. Mamkaeva from samples collected in April 2014, from a roadside ditch with abundant leaf litter in the vicinity of Sergievka park, Petershof, St. Petersburg (X-113), around the Opolie village, Kingisepp District, Leningrad Province (X-114) and from the Narvskiy reservoir, Kingisepp District, Leningrad Province (X-130), Russia. The strains were maintained in culture on Tribonema gayanum (strain 20 CALU) as described in Karpov et al. (2016). Infected strains (enrichment cultures) are available upon request.
Light and transmission electron microscopy
Light and DIC microscopy observations of living cultures were carried out on a Zeiss Axioplan microscope equipped with black and white MRm Axiocam. For electron microscopy, the algal filaments were fixed with 2% glutaraldehyde on 0.1M cacodylate buffer for two hours on ice, rinsed in the same buffer, and fixed with 1% OsO4 during one hour on ice. After rinsing in 0.1M cacodylate buffer for 5 min, the filaments were dehydrated in alcohol series and in propylen oxide, then embedded in Spurr resin (Sigma Aldrich, St Louis, USA). Ultrathin sections were prepared using a Leica Ultracut ultratome with a diamond knife. After double staining, the sections wereobserved using a JEM 1400 (Jeol) microscope equipped with digital camera Olympus Veleta.
Amplification and sequencing of Amoeboradix gromovi 18S and 28S rRNA genes
18S rRNA genes were amplified from mixed cultures of strains X-113 (K1 culture), X-114 (K2 culture) and X-130 (K3 culture) using combinations of the forward eukaryote-specific 82F (5’-GAAACTGCGAATGGCTC) (López-García et al, 2001) and fungi-specific NS1 (5’- GTAGTCATATGCTTGTCTC) (Vainio and Hantula, 2000) primers and the reverse eukaryote-specific 1520R (5’-CYGCAGGTTCACCTAC) (López-García et al. 2001) and fungi-specific AU4 (5’- RTCTCACTAAGCCATTC) (Vandenkoornhuyse et al. 2002) primers. Amplified genes were cloned using TOPO TA Cloning System (Thermo Fisher Scientific, MA, USA) and sequenced using vector primers (Genewiz, Takeley, UK). Several eukaryotic genes belonging to various taxa were identified in the cultures (not shown). In order to ascertain the molecular identity of A. gromovi in those mixed cultures, we isolated single sporangia of strains X-113 and X-114 using an Eppendorf TransferMan 4r micromanipulator system mounted on an inverted microscope Leica DMI3000 B equipped with a Leica DFC450 camera. We subsequently amplified the 18S + 28S rDNA cluster from single sporangia using the primer combination NS1 and 28S-4R (5’- TTCTGACTTAGAGGCGTTCAG) (D. Moreira, pers. com.) followed by semi-nested PCR amplifications covering different, but overlapping, rDNA cluster regions with primers NS1 + 1909R (5’- AGAGAGTCATAGTTACTCC) (D. Moreira, pers. com.) and 1611F (5’-CCGCAKCAGGT TCCAAG) (D. Moreira, pers. com.) + 28S-4R. The cloned cluster regions were sequenced using PCR primers and several internal primers and assembled using CodonCode Aligner (http://www.codoncode.com/aligner/). Finally, to further validate the identity of the zoospores, we micromanipulated individual amoeboid zoospores of strain X-113, and we amplified the 18S rRNA gene using fungal-specific primers NS1 + AU4. The 18S rRNA sequence obtained from zoospores was identical to that of X-113 and X-114 18S+28S rDNA clusters sporangia and to previously sequenced 18S rRNA genes from K1 and K2 cultures. The sequence of the nearly full gene cluster containing the 18S, 5.8S and 28S rRNA genes of Amoeboradix gromovi strain X-113 has been deposited at GenBank with accession number MG256970.
Molecular phylogenetic analyses
18S and 28S rRNA gene sequences from a large diversity of fungi and several other opisthokont species used as outgroup (see Supplementary Fig. S3 and Table S1) were retrieved from the Silva database (https://www.arb-silva.de/). These sequences were included in two independent alignments, one for the 18S and one for the 28S rRNA gene, after adding the corresponding A gromovi sequences. Multiple sequence alignments were carried out using MAFFT (Katoh et al. 2002) and the alignments were trimmed to remove ambiguously aligned regions with trimAl (Capella-Gutierrez et al. 2009). The trimmed 18S and 28S rRNA gene sequence of each species were then concatenated with an in-house Python script (provided as supplementary file). The concatenated dataset (3761 nucleotides) was used for phylogenetic reconstruction. Bayesian inference analyses were done with 4 chains using PhyloBayes (Lartillot et al. 2013) with the CAT-GTR model and a length of 80,000 cycles. Convergence between the 4 chains was estimated by calculating the difference in frequency for all their bipartitions using a threshold maxdiff <0.1. The first 20,000 points were discarded as burn-in. The posterior consensus was computed by selecting 1 tree every 10 over the 4 chains.
Supplementary Material
Acknowledgments
The morphological study of strain X-113, molecular phylogeny of strain X-130, and the manuscript writing have been supported by the RSF grant no. 16-14-10302. The study of molecular phylogeny of strains X-113 and X-114 and the manuscript writing have been supported by the ERC Advanced Grant No. 322669 ‘ProtistWorld’. SK thanks to scholarship program "Jean d'Alembert" of Paris-Saclay University, French Agence Nationale de la Recherche (project ANR-15-CE32-0003 «ANCESSTRAM»), and the program of the Russian Academy of Sciences "Evolution of the biosphere". The investigated strains have been cultivated at the Center for Culturing Collection of Microorganisms of Research park of St. Petersburg State University. We thank the Research Resource Center for Molecular and Cell Technologies (RRC MCT) at St. Petersburg State University (SPbSU) for access to the EM facilities and the UNICELL single cell genomics facility at the CNRS-Université Paris-Sud. We thank the editor and two anonymous reviewers for insightful comments.
Abbreviations
- aw
algal wall
- c
centriole
- ch
chloroplast
- cv
contractile vacuole
- cw
cyst wall
- cy
cyst
- cz
centriolar zone
- f
filopodia
- Ga
Golgi apparatus
- h
host
- hb
hub with spokes at the base of kinetosome and centriole
- hc
heterochromatin
- k
kinetosome
- l
lipid globule
- m
mitochondrium
- mi
microbody
- mt
microtubules
- n
nucleus
- nu
nucleolus
- pa
papilla
- pl
plasma membrane
- pc
pseudocilium
- rh
rhizoid
- sp
sporangium
- sw
sporangium wall
- v
vesicles
- vd
vesicles with degraded material
- vp
viral particle
- zo
zoospores
References
- Barr D. Taxonomy and phylogeny of chytrids. BioSystems. 1978;10:153–165. doi: 10.1016/0303-2647(78)90037-0. [DOI] [PubMed] [Google Scholar]
- Barr D. Evolution and Kingdoms of organisms from the perspective of mycologist. Mycologia. 1992;84:1–11. [Google Scholar]
- Barr D, Allan P. Zoospore ultrastructure in Polymyxa graminis (Plasmodiophoromycetes) Can J Bot. 1982;60:2496–2504. [Google Scholar]
- Braun A. Uber einige neue Arten der Gattung Chytridium und die damit verwandte Gattung Rhizidium. Monatsber Berlin Akad. 1856:587–592. [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch JN. New or little known Chytridiales. Mycologia. 1935;27:160–175. doi: 10.2307/3754051. [DOI] [Google Scholar]
- Gromov BV, Titova NN. Catalogue of Microbial Cultures in the Collections of the USSR. RAS; Moscow: 1991. CALU-Collection of algal cultures in the laboratory of microbiology of Biological Institute of Sankt-Petersburg University; pp. 76–125. [Google Scholar]
- Guichard P, Hachet V, Majubu N, Neves A, Demurtas D, Olieric N, Fluckiger I, Yamada A, Kihara K, Nishida Y, Moriya S, et al. Native Architecture of the Centriole Proximal Region Reveals Features Underlying Its 9-Fold Radial Symmetry. Current Biology. 2013;23:1620–1628. doi: 10.1016/j.cub.2013.06.061. [DOI] [PubMed] [Google Scholar]
- James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota) Mycologia. 2006;98:860–871. doi: 10.3852/mycologia.98.6.860. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karling JS. Chytridiomycetarum Iconographia. J. Cramer; Vaduz: 1977. p. 414. [Google Scholar]
- Karpov SA. Structure of the axoneme and flagellar appendages in algae and colourless flagellates. Botan Zhurn. 1987;72:3–14. (in Russian) [Google Scholar]
- Karpov SA, Mikhailov KV, Mirzaeva GS, Mirabdullaev IM, Mamkaeva KA, Titova NN, Aleoshin VV. Obligately Phagotrophic Aphelids Turned out to Branch with the Earliest-diverging Fungi. Protist. 2013;164:195–205. doi: 10.1016/j.protis.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Karpov SA, Kersanach R, Williams DM. Ultrastructure and 18S RNA gene sequence of a small heterotrophic flagellate Siluania monomastiga gen. et sp. nov. (Bicosoecida) Europ J Protistol. 1998;34:415–425. [Google Scholar]
- Karpov SA, Kobzeva AA, Mamkaeva MA, Mamkaeva KA, Mikhailov KV, Mirzaeva GS, Aeoshin VV. Gromochytrium mamkaevae gen. et sp. nov. and two new orders: Gromochytriales and Mesochytriales (Chytridiomycetes) Persoonia. 2014;32:115–126. doi: 10.3767/003158514X680234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov SA, Mamanazarova KS, Popova OV, Aleoshin VV, James TY, Mamkaeva MA, Tcvetkova VS, Vishnyakov AE, Longcore JE. Monoblepharidomycetes diversity includes new parasitic and saprotrophic species with highly intronized rDNA. Fungal Biol. 2017;121:729–741. doi: 10.1016/j.funbio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Karpov SA, Tcvetkova VS, Mamkaeva MA, Torruella G, Timpano H, Moreira D, Mamanazarova KS, López-García P. Morphological and Genetic Diversity of Opisthosporidia: New Aphelid Paraphelidium tribonemae Gen. et Sp. Nov. J Eukaryot Microbiol. 2016;64:204–212. doi: 10.1111/jeu.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Letcher PM, Powell MJ. A taxonomic summary and revision of Rhizophydium (Rhizophydiales, Chytridiomycota) Alabama University Printing; Tuscaloosa, USA: 2012. No. 1. Imprint. [Google Scholar]
- Letcher PM, Lee PA, Lopez S, Burnett M, McBride RC, Powell MJ. An ultrastructural study of Paraphysoderma sedebokerense (Blastocladiomycota), an epibiotic parasite of microalgae. Fungal Biology. 2016;120:324–37. doi: 10.1016/j.funbio.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Letcher PM, Powell MJ, Churchill PF, Chambers JG. Ultrastructural and molecular phylogenetic delineation of a new order, the Rhizophydiales. Mycological Research. 2006;110:898–915. doi: 10.1016/j.mycres.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Longcore JE, Simmons DR. The Polychytriales ord. nov. contains chitinophilic members of the rhizophlyctoid alliance. Mycologia. 2012;104:276–294. doi: 10.3852/11-193. [DOI] [PubMed] [Google Scholar]
- López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Mamkaeva KA, Pljusch AV, Mamkaeva MA, Karpov SA. Amoeboradix gromovi gen. et sp. nov. - enigmatic parasite of the alga Tribonema gayanum. Protistology. 2007;5:53. [Google Scholar]
- Mozley-Standridge SE, Letcher PM, Longcore JE, Porter D, Simmons DR. Cladochytriales – a new order in Chytridiomycota. Mycological Research. 2009;113:498–507. doi: 10.1016/j.mycres.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Powell MJ. Chytridiomycota. In: Archibald, Simpson, Slamovits, Margulis, Melkonian, Chapman, Corliss, editors. Handbook of the Protists. Springer International Publishing; 2016a. pp. 1–36. [Google Scholar]
- Powell MJ. Blastocladiomycota. In: Archibald, Simpson, Slamovits, Margulis, Melkonian, Chapman, Corliss, editors. Handbook of the Protists. Springer International Publishing; 2016b. pp. 1–25. [Google Scholar]
- Schrevel J, Goldstein S, Kuriyama R, Desportes I. Biology and cell organization of gregarines during gamogony. In: Desportes I, Schrevel J, editors. Treatise on Zoology – Anatomy, Taxonomy, Biology. The Gregarines. Vol. 1. 2013. pp. 110–136. [Google Scholar]
- Sekimoto S, Rochon A, Long JE, Dee JM, Berbee ML. A multigene phylogeny of Olpidium and its implications for early fungal evolution. BMC Evolutionary Biology. 2011;11:331. doi: 10.1186/1471-2148-11-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seravin LN. Variations in the axoneme constitution of flagella and the origin of this organelle. Cytologiya. 1985;27:971–985. (in Russian) [Google Scholar]
- Simmons DR, James TY, Meyer AF, Longcore JE. Lobulomycetales, a new order in the Chytridiomycota. Mycological Research. 2009;113:450–460. doi: 10.1016/j.mycres.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, Bernard C, Fenchel T, Patterson DJ. The organization of Mastigamoeba schizophrenia n. sp.: more evidence of ultrastructural idiosyncrasy and simplicity in pelobiont protists. Europ J Protistol. 1997;33:87–98. [Google Scholar]
- Sparrow FK. Inoperculate chytridiaceous organisms collected in the vicinity of Ithaca, N.Y., with notes on other aquatic fungi. Mycologia. 1933;25:513–535. doi: 10.2307/3754109. [DOI] [Google Scholar]
- Sparrow FK. Aquatic Phycomycetes. The University of Michigan Press; Ann Arbor: 1960. [Google Scholar]
- Vainio E, Hantula J. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycological Research. 2000;104:927–936. [Google Scholar]
- Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JP. Extensive fungal diversity in plant roots. Science. 2002;295:2051. doi: 10.1126/science.295.5562.2051. [DOI] [PubMed] [Google Scholar]
- Zopf W. Zur Kenntniss der Phycomyceten. I. Zur Morphologie und Biologie der Ancylisteen und Chytridiaceen. Nova Acta Acad Leop-Carol. 1884;47:143–236. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.