Figure 1.
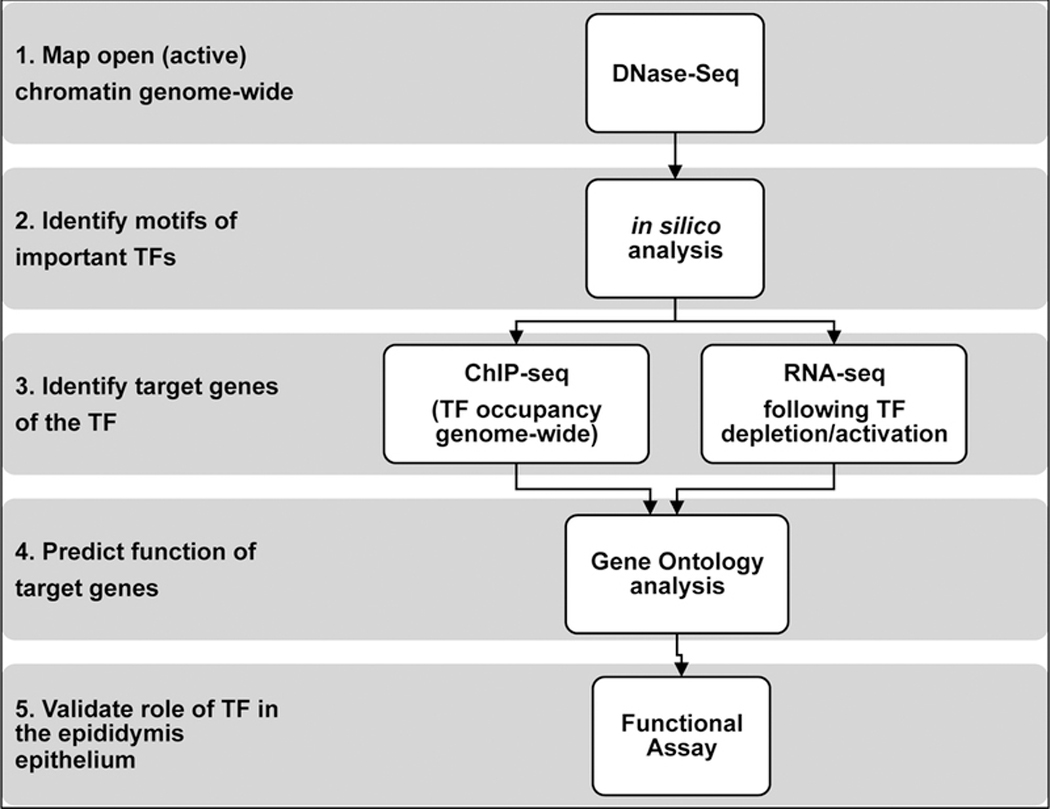
Combining different NGS-based experiments to build transcriptional regulatory networks in the epididymis epithelium. 1. DNase-seq maps open (active) regions of chromatin to identify potential regulatory elements. 2. ChIP-seq identifies transcription factor occupancy genome-wide and its target genes. 3. RNA-seq following TF activation/depletion determines the contribution of the TF to the transcriptome. 4. Gene ontology analysis to predict the function of target genes. 5. Validate identified role of TF with a functional assay.
