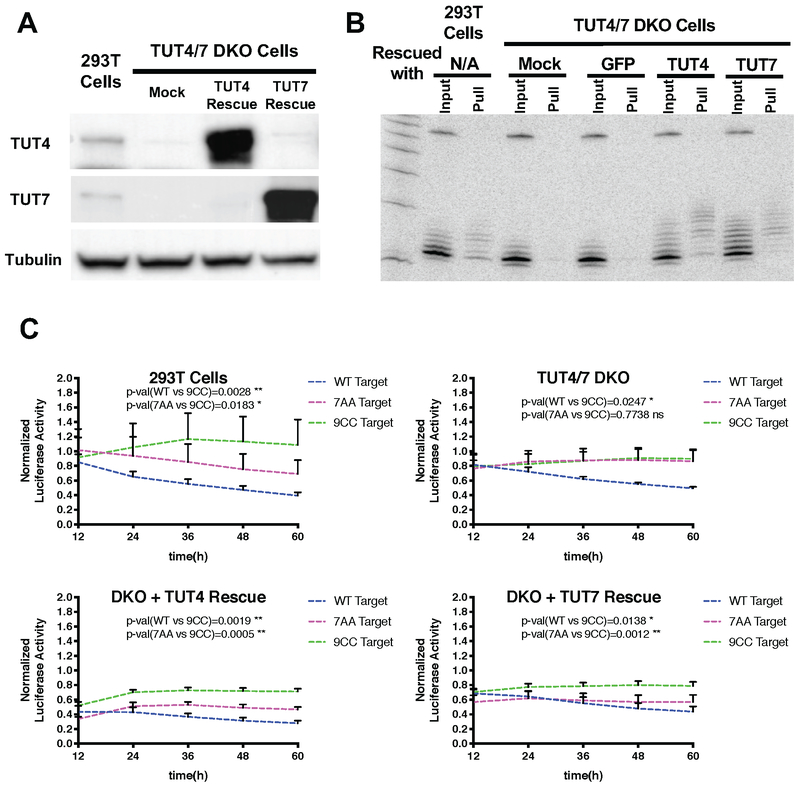
Figure 4. TUT4/TUT7-dependent repression of non-canonical targets.
(A) TUT4 and TUT7 double knockout (DKO) cells were generated by CRISPR/Cas9 in HEK293T cells. Western blot was used to assess the expression of TUT4 and TUT7 proteins. Tubulin was detected and served as a loading control (B) Small RNAs associated with 7AA targets were detected by the biotin-pull-down assay performed in HEK293T cells as well as in TUT4/7 DKO cells with or without TUT4/7 rescue. (C) Various reporters were each coexpressed with miR-27a in HEK293T cells and TUT4/7 DKO cells with or without TUT4/7 rescue. Gaussia luciferase activities were first normalized with firefly luciferase activity from a co-transfected plasmid. Then the percentage of normalized Gaussia luciferase activity compared to the negative control treated with a non-targeting control miRNA was shown in the figure. The value of negative control was designated as 1. Error bars represent the standard deviation from three biological replicates. See also Figure S4.