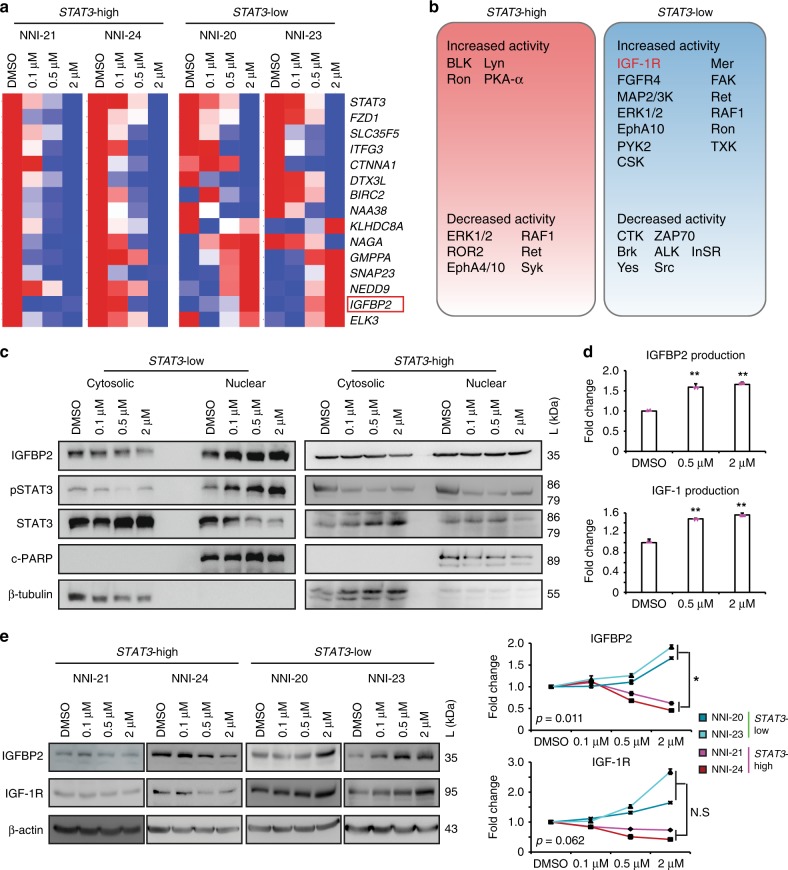
Fig. 3.
Mechanistic gene candidates identified by NNI-STAT3 gene signature. a Winnowed gene list across patient tumors identified candidates uniquely upregulated in STAT3-high tumors. A dose-dependent differential gene expression after signal transducers and activators of transcription 3 (STAT3) inhibitor AZD1480 treatment distinguished cooperative genes responsible in the STAT3-resistant profile. Results are mean of triplicate experiments. b Graphical illustration of responsive and resistant protein tyrosine kinase candidates in STAT3-high and -low cell lines treated with AZD1480 using computational workflow described in Supplementary Fig. 4c. c Treatment of cells with STAT3 inhibitor (AZD1480) demonstrated an increase in pSTAT3 and insulin-like growth factor binding protein 2 (IGFBP2) expression levels in the nuclear fraction of STAT3-low cells but not in STAT3-high. d Fold change differences in secreted proteins demonstrated increased IGFBP2 and insulin-like growth factor 1 receptor (IGF-1R) in resistant cells post-treatment with STAT3 inhibitor. Results are mean of triplicate experiments. e STAT3-high glioblastoma (GBM) cells displayed modest reduction in IGF-1R and IGFBP2 expression levels. In contrast, IGF-1R and IGFBP2 protein expression in STAT3-low cells increased dose-dependently upon AZD1480 treatment, albeit IGF-1R was marginally insignificant. Fold change differences in protein expression of IGFBP2 and IGF-1R were compared between STAT3-high and -low GBM cells. *p < 0.05; **p < 0.01; STAT3-high versus STAT3-low. For statistical analysis, two-sided Student’s t test was used. Error bars represent standard deviation of the mean