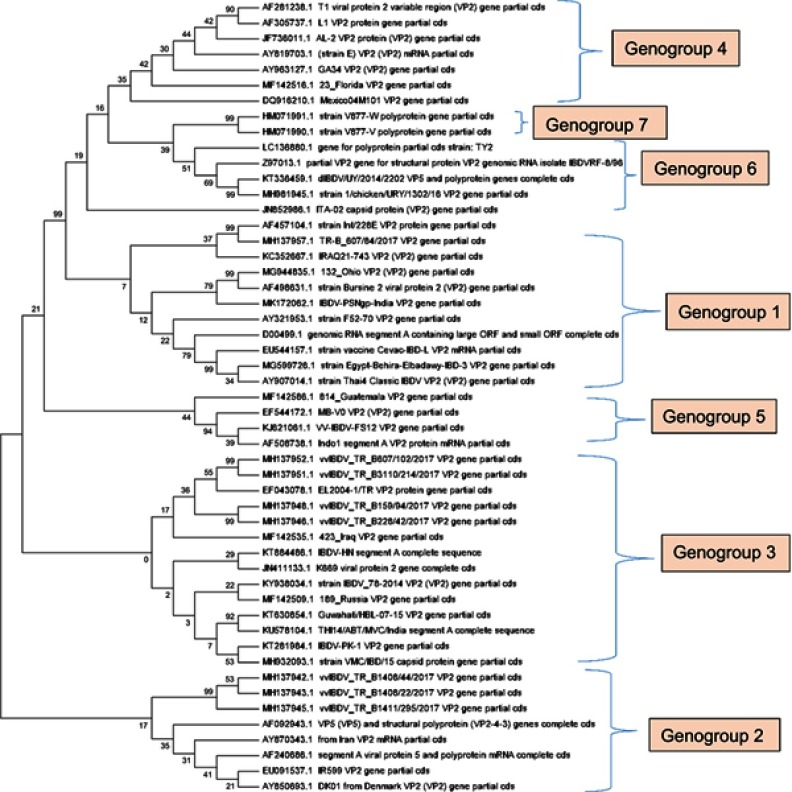
Figure 1.
Phylogenetic analysis of the nucleotide sequences of hVP2 infectious bursal disease virus (IBDV). The evolutionary history was inferred using the Neighbour-Joining method. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The analysis involved 51 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated. There were a total of 336 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.90