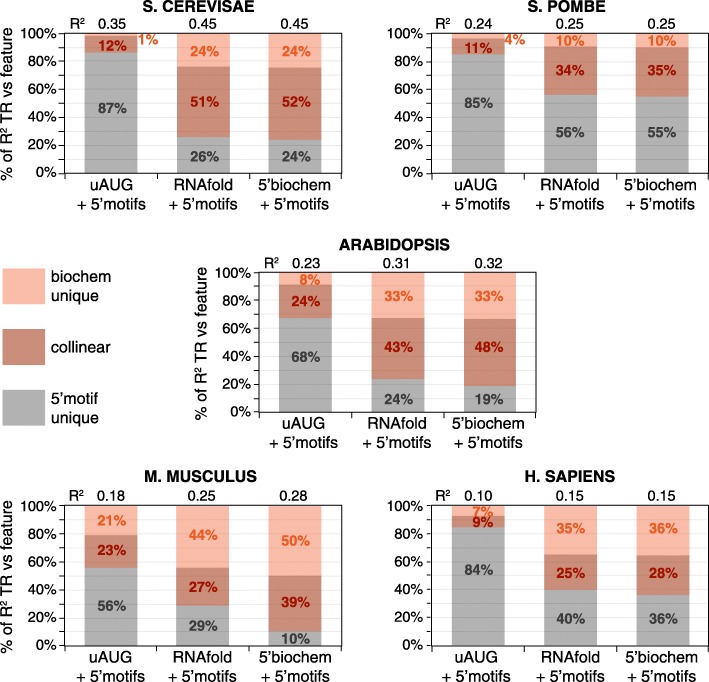
Fig. 7.
Control by “RNAfold” and “uAUG” is collinear with that by “5′motifs.” Two 5′ features each partially capture control by a biochemical process: “uAUG,” repression due to translation of uORFs; and “RNAfold,” inhibition by RNA secondary structure. A third feature, “5′motifs,” captures 5′ sequences that correlate with translation, without regard to biochemical mechanism. To determine how similar information captured by “5′motifs” is to that in “uAUG” and “RNAfold,” three multivariate models were constructed that combined “5′motifs” either with “uAUG,” with “RNAfold,” or with a third feature (5′biochem) that combines both “uAUG” and “RNAfold.” The R2 coefficients of these feature sets vs TR are shown (top). The percent of the R2 values contributed by “5′motifs” (gray) or the biochemical process(es) (orange) is shown by length along the y-axis. Because some of this information is collinear, the R2 coefficient of the combined models is less than that of the sum of the individual contributions. This collinear portion is shown by the darker orange shading, while the lighter orange and the gray shading show the unique contributions